Assessment of the Effects of IL9, IL9R, IL17A, and IL17F Gene Polymorphisms on Women with Allergic Rhinitis in Shahrekord, Iran
- *Corresponding Author:
- Dr. Amiri M
Department of Epidemiology and Biostatistics, School of Health, Shahrekord University of Medical Sciences, Shahrekrod, Iran.
E-mail: masoud.amiri@yahoo.com
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Citation Fatahi F, Chaleshtori A, Samani KG, Mousavi SM, Zandi F, Heydari S, et al. Assessment of the effects of IL9, IL9R, IL17A, and IL17F gene polymorphisms on women with allergic rhinitis in Shahrekord, Iran. Ann Med Health Sci Res 2016;6:216-23.
Abstract
Background: The genes encoding IL9, IL9R, IL17A, and IL17F have recently been implicated in the genetic basis of rhinitis and allergy. Aim: The purpose of this study was to assess the association of the single nucleotide polymorphisms (SNPs) of IL9, IL9R, IL17A, and IL17F and potential interaction of these genes with the determination of IgE levels in women with allergic rhinitis (AR) in Shahrekord, Iran. Subjects and Methods: In a case–control study, SNPs from the IL9, IL9R, IL17A, and IL17F were genotyped in 394 random samples including 195 AR patients and 199 normal controls. Enzyme‑linked immunosorbent assay was performed for the determination of serum total IgE levels. The Student’s t‑test was used to compare the differences. The Chi‑square test was performed to compare proportions of cases with different clinical features among cases with different genotypes. The genotype and allele frequencies were obtained by direct counting. Hardy–Weinberg equilibrium was tested between cases and controls separately. The relative risk associated with rare alleles was estimated as an odds ratio with 95% confidence interval. P ≤ 0.05 was considered statistically significant. Results: The rs731476 SNP in the IL9R was significantly associated with the AR phenotype in women. No association was found between any of the other SNPs in IL9, IL17A, and IL17F genes and AR. In the gene–gene interaction analysis, we found that IL9R/IL9 genotype rs731476 T‑/rs2069885 G conferred a higher risk for AR phenotype development. We also did not find a significant association in terms of IgE levels between cases and controls. Conclusion: Our result suggests that the rs731476 SNP located in the IL9R is associated with an increased susceptibility to AR in females. In a subsequent gene–gene interaction analysis, the rs731476 T‑/rs2069885 G‑genotype combination (IL9R/IL9) has significantly been associated with the development of the AR phenotype
Keywords
Allergic rhinitis, Gene–gene interaction, IL17A, IL17F, IL9, IL9R, Single nucleotide polymorphisms
Introduction
Allergic rhinitis (AR), which is among the most common allergic disorders, is an inflammatory disorder of the nasal mucosa caused by an IgE-mediated response, following exposure to an allergen. The clinical characteristics of AR include itching, sneezing, rhinorrhea, and nasal congestion or stuffiness that can be reversible either spontaneously or by treatment.[1] With a prevalence of 9–42 among the general population (and 10–15% in Iran), AR is considered as a global healthcare problem.[2] It has a considerable impact on quality of life which is negatively affected patients’ social life and school performance. AR is also a risk factor for the development of asthma. The exact etiology of AR is currently unknown but could involve a complex interaction between genetic predisposition and environmental exposure to different factors including allergens. Although the disease does have a hereditary component with a 45–60% concordance among monozygotic twins and an estimated 0.33–0.75 of heritability, it does not follow a certain Mendelian hereditary pattern and is widely considered to be complex. Thus, as expected, an array of genetic, epigenetic, and environmental factors should be engaged.[3,4]
The interleukin-9 receptor gene (IL9R), belonging to the hematopoietin receptor superfamily, is located on the pseudoautosomal region of X and Y chromosomes (Xq28 and Yq12).[5] It is expressed on T cells, macrophages, mast cells, eosinophils, and neutrophils.[6] Upon IL9 binding to IL9R, JAK-STAT signaling pathway is activated.[7]
In humans, IL9 is located on chromosome 5 (5q31–35) between IL3 gene and early growth response-1.[8] Activated IL9 receptor plays a key role in immunologic processes such as T cell development.[9] It is also involved in the prevention of apoptosis.[10] It can prompt the release of chemotactic factors from bronchial epithelial cells and smooth muscle cells.[11,12] IL9/IL9R can act directly on B lymphocytes and regulate IgE synthesis. Therefore, it may play a major role in the development of allergy.
The IL17 and IL17F genes, mapped on the same chromosome at position 6p12, share a 50%, homology. These genes are expressed by the activated T cells which, in turn, induce the expression of cytokines and chemokines.[13] The available evidence suggests that IL17F is an excellent candidate gene for chronic inflammatory disease including ulcerative colitis,[14] asthma,[15] inflammatory bowel disease,[16] and AR.[17] IL17F is a powerful pro-inflammatory cytokine capable of inducing other cytokines.[18] Thus, it may be important for neutrophilic inflammation in acute airway inflammation[19-21] and AR.[17] There are some reports about the association of single nucleotide polymorphisms (SNPs) with increased risk of AR as discussed below. In this study, we performed an association study on case and control populations to evaluate the possible association of SNPs from IL9 (rs2069885), IL9R (rs731476),IL17A (rs2275913), and IL17F (rs763780) genes, both alone and in combination, with an increased risk of AR.
Subjects and Methods
In a case–control study, AR samples were obtained from nonasthmatic AR patients examined at Kashani Hospital, an educational hospital in Shahrekord University of Medical Science, Shahrekord, Iran, between 2009 and 2010. All samples were selected from Fars ethnic group randomly. Patient’s written informed consent was obtained.
We selected women because (1) women would present the allergic-related diseases more than men; (2) the studied genes in IL9 were located on sexual chromosomes; and (3) there are few studies on women than men in this regard. In this study, SNPs from the IL9, IL9R, IL17A, and IL17F were genotyped in 394 random samples including 195 AR patients and 199 normal controls. Enzyme-linked immunosorbent assay (ELISA) was performed for determination of serum total IgE levels. The systematic random sampling was used to select cases and controls using the file numbers. The controls were selected from patients who were referred but did not have AR. The total numbers of cases and controls were obtained using Cochran’s sample size formula. Based on the discussion with experts, the questionnaire did not need to verify for reliability and validity due its questions which were only related to general demographic and history questions.
The diagnosis criteria for AR included positive history of disease, positive physical examination (turbinate hypertrophy and submucosal inflammation), ruling out of other causes of nasal obstruction, and not having anatomic disorders. Among those with asthma or atopic dermatitis, patients who might have got AR as secondary symptom were excluded from the study. Patients with positive family history (having at least three persons with AR phenotype in their family) were selected for the study. In total, 195 patients with AR and 199 healthy controls (all women) were included in the study. The normal cases were recruited locally, primarily from medical students and volunteers with no prior history of autoimmune or inflammatory diseases such as systemic lupus erythematosus, rheumatoid arthritis or inflammatory bowel disease, AR, or asthma. They were matched in terms of gender, ethnicity, and age (with a range of 5-year) with the patient group. All the cases with anatomic problem in their nose, related to cancer, and smokers were also excluded from the study. This study was approved by the Ethics Committee of Shahrekord University of Medical Sciences and was conducted according to the declaration of Helsinki principles. Written informed consents were obtained from all the participants.
Blood samples were drawn by venipuncture in EDTA-containing tubes. Serum total IgE levels were determined by human IgE ELISA kit (BioCheck, USA). A serum IgE level was considered elevated if it exceeded the highest reference value of 150 IU/ml.
DNA extraction and genotyping
Genomic DNA was extracted using a standard phenol/ chloroform extraction method. IL9 rs2069885, IL9R rs731476, IL17A rs2275913, and IL17F rs763780 genotyping were performed by polymerase chain reaction (PCR)-restriction fragment length polymorphism. Primer sequences for IL17A G-152A and IL17F 161His-Arg were as follows:
Forward: 5’-CAGAAGACCTACATGTTACT-3’, Reverse: 5’-GTAGCGCTATCGTCTCTCT-3’ for IL17A
Forward: 5’-GTTCCCATCCAGCAAGAGAC-3’, Reverse 5’-AGCTGGGAATGCAAACAAAC-3’ for IL17F
Forward: 5’CATCATTTGAGTCACTCTGTCCTT-3’, Reverse: 5’-TTGCCTCTCATCCCTCTCAT-3’ for IL9.
The primers for IL9R were as follows:
Forward: 5-CTTGTCCACCCAACACCTCT-3, Reverse: 5-CTGCATCCGTGAGGTAAAGG-3.
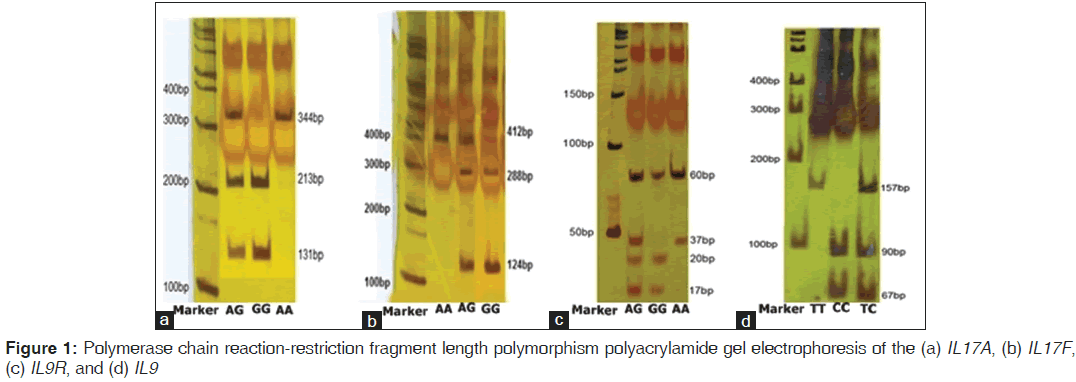
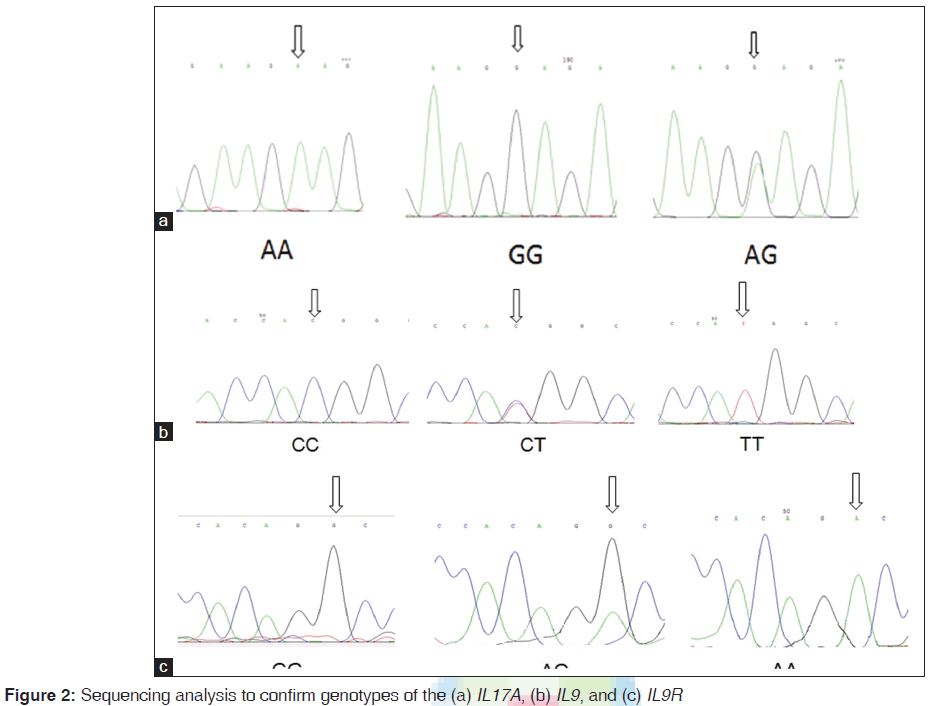
The reverse primer was designed in such a way to create a new restriction digest site for a restriction enzyme. Primers were designed by primer3.[22] The PCR amplification was performed in a total volume of 25 μl mixture containing: 100-ng genomic DNA, 1.0 μM of each primer, 200 μM of each dNTP, 2.0 μM of MgCl2 and 1.0 U Taq DNA polymerase and 10 μl Taq buffer (Fermentas, Germany) using the Astec gradient 96 (Astec, Japan). PCR products were digested overnight at 37°C with NlaIII (Fermentas, Germany) for IL17F 161His-Arg and IL9Rs2069885, XmnI for IL17A rs2275913, and HaeIII for IL9Rrs731476 genotypes. They were resolved on 10% polyacrylamide gel electrophoresis for IL9Rrs731476. PCR products were shown to be digested into three fragments for all of them except IL9R that digested in four fragments [Figure 1]. To confirm the genotyping results, randomly selected PCR samples were tested by DNA sequencing [Figure 2].
Determination of levels of IgE in cases and controls
The levels of IgE were determined using specific ELISA kits (Ray Biotech Inc., Norcross, Georgia, USA).
Statistical methods
The quantitative variables were expressed as mean (standard deviation). The Student’s t-test was used to compare the differences. The Chi-square test was performed to compare proportions of cases with different clinical features among cases with different genotypes. The genotype and allele frequencies were obtained by direct counting. Hardy–Weinberg equilibrium was tested between cases and controls separately. The relative risk associated with rare alleles was estimated as an odds ratio (OR) with 95% confidence interval (CI). P ≤ 0.05 was considered statistically significant. All statistical analyses were performed using SPSS version 17.0 (SPSS Inc., Chicago, IL, USA).
To determine whether interactions between the rs731476SNP, rs2069885 SNP, rs2275913 SNP, and rs763780 SNP affect disease susceptibility, association test was conducted using genotypic combinations of these four SNPs. A logistic regression model was used to evaluate the statistical significance, using age and gender as adjusting covariates. The ORs for each genotypic combination were also obtained from the logistic regression analysis, after the genotypes were subdivided into two groups for each locus (GG and others for rs731476, TT and others for rs2069885, AA and others for rs2275913, and GG and others for rs763780).
Results
The frequencies of the genotypes
A total of four SNPs from the IL9, IL9R, IL17A, and IL17F were genotyped from the 394 samples. Information from the SNPs is shown in Table 1, including genomic function, chromosomal position, dbSNP ID, and minor allele frequency. The Fisher exact test indicated that all genotyped SNPs were in Hardy–Weinberg equilibrium. The distributions of the allelic and genotypic frequencies for the four SNPs were compared among the AR and normal groups. The allelic and genotypic frequencies of these four SNPs are shown in Table 2. The SNP rs731476 from the IL9R was genotyped, showing significantly different allelic and genotypic distributions between the AR and normal groups (allelic P = 0.01; genotypic P < 0.001). For the allelic test, the resulting OR was 0.5 (95% CI = 0.34–0.77) [Table 2]. No significant allelic or genotypic associations were found between AR and any of the three SNPs from the IL9, IL17A, and IL17F or the rs2069885, rs2275913, and rs763780 SNPs. Polymorphisms in the four genes were also compared with total IgE level between cases and controls to identify any significant correlation with AR; as a result, the SNPs in the IL9, IL9R, IL17A, and IL17F genes were observed to have significant associations with these variables.
| Gene | rs number | Chromosomes | Function | MAF* |
|---|---|---|---|---|
| IL9 | rs2069885 | Chromosomes 5 | Exon 5 | 0.1436 |
| IL9R | rs731476 | Chromosomes X/Y | Intron 1 | 4974 |
| IL17A | rs2275913 | Chromosomes 6 | Promoter | 0.3538 |
| IL17F | rs763780 | Chromosomes 6 | Exon 3 | 0.154 |
Table 1: Single nucleotide polymorphism markers genotyped for the case-control samples (dbSNP build 126)
| Group a | Allele | OR (95% CI) | P | ||
|---|---|---|---|---|---|
| A | G | ||||
| NR | 132 (0.6633) | 67 (0.4974) | 0.51 (0.34-0.77) | 0.01 | |
| AR | 98 (0.5026) | 97 (0.4974) | |||
| Genotype | |||||
| AA | AG | GG | |||
| NR | 66 (0.3317) | 133 (0.6683) | 0 | <0.001 | |
| AR | 48 (0.2462) | 100 (0.5128) | 47 (0.2410) | ||
| Group b | Allele | ||||
| C | T | ||||
| NR | 27 (0.1357) | 172 (0.8643) | 1.07 (0.60-1.89) | 0.82 | |
| AR | 28 (0.1436) | 167 (0.8564) | |||
| Genotype | |||||
| CC | CT | TT | |||
| NR | 0 | 53 (0.2663) | 146 (0.7373) | 0.12 | |
| AR | 4 (0.02051) | 47 (0.2411) | 144 (0.7385) | ||
| Group c | Allele | ||||
| A | G | ||||
| NR | 76 (0.3819) | 123 (0.6181) | 0.89 (.59-1.53) | 0.56 | |
| AR | 69 (0.3538) | 126 (0.6462) | |||
| Genotype | |||||
| AA | AG | GG | |||
| NR | 20 (0.1005) | 112 (0.5628) | 67 (0.3369) | 0.70 | |
| AR | 24 (0.1231) | 89 (0.4564) | 82 (0.4205) | ||
| Group d | Allele | ||||
| A | G | ||||
| NR | 188 (0.9447) | 11 (0.0553) | 0.89 (0.38-2.07) | 0.79 | |
| AR | 183 (0.9385) | 12 (0.6154) | |||
| Genotype | |||||
| AA | AG | GG | |||
| NR | 177 (0.8894) | 22 (0.1106) | 0 | 0.70 | |
| AR | 171 (0.8769) | 24 (0.1231) | 0 |
Table 2: Comparison of allelic and genotypic frequencies between the two allergic rhinitis and normal control groups: (a) rs731476 single nucleotide polymorphism from the IL9R gene, (b) rs2069885 single nucleotide polymorphism from the IL9 gene, (c) rs2275913 single nucleotide polymorphism from the IL17A gene, and (d) rs763780 single nucleotide polymorphism from the IL17F gene. Age and gender were used as adjusting covariates
Gene–gene interaction between IL9 and IL9R, IL17A, and IL17F
To investigate whether interactions between the rs2069885 and rs731476 SNPs and between rs2275913 and rs763780 affect the disease susceptibility, an association test was conducted using genotype combinations of each of the two SNPs. After analysis, these data showed that two SNP genotype combinations are significantly increased the risk of AR. When interaction between an allele from IL9 and G allele from IL9R was compared between the two AR and normal controls, the resulting OR was 0.67 (95% CI = 0.46–0.98) (P = 0.04). These results suggest that the combination of the two genotypes from other two genes has no effect on increasing the disease susceptibility [Table 3].
| Interactions | AR versus NR OR (95% CI) | P |
|---|---|---|
| rs731476/rs2069885 | ||
| A-/C- | 1.40 (0.77-2.52) | 0.13 |
| G-/C- | 0.72 (0.39-1.32) | 0.29 |
| T-/A- | 1.31 (0.93-1.83) | 0.12 |
| G-/T- | 0.67 (0.46-0.98) | 0.04 |
| rs2275913/rs763780 | ||
| A-/A- | 1.07 (0.73-1.57) | 0.72 |
| A-/G- | 0.83 (0.34-2.01) | 0.68 |
| G-/A- | 1.05 (0.76-1.45) | 0.75 |
| G-/G- | 0.94 (0.40-2.21) | 0.88 |
Table 3: Gene-gene interaction IL9 and IL9R, IL17A, and IL17F
Discussion
AR is largely considered to be a complex disease with poorly understood genetics. Although many genes have been studied, few have been successfully associated with the disease. In the present investigation on four different autoimmune-related genes which were chosen on the basis of candidate gene approach, for the first time, the rs731476 SNP in IL9R was found to be significantly associated with AR in the studied women. In addition, the rs731476 T-/rs2069885 G-combination in the IL9/IL9R genes was found significantly increase the risk of developing the AR phenotype (OR = 0.67; 95% CI = 0.46–0.98; P = 0.04).
Allelic association between IL9 and total serum IgE levels has been previously demonstrated.[23] SNPs from IL9, IL9R, IL17A, and IL17F and total serum IgE, in cases (n = 195) and controls (n = 199), have not been correlated (P = 0.08). Both IL9 and IL9R have been implicated in asthma pathogenesis.[23-26] Although, according to a recent study, IL9 is produced in larger quantities by Treg than Th2 cells,[27] most of the present evidence indicates that the physiological function of IL9 is somehow connected to the Th2 signaling – IL9 is involved in defense against helminthic infections; it also contributes to allergic reactions in the lung.[28] Furthermore, IL9 may mediate the activity of both T cells and Treg cells.[29] IL9R gene polymorphism has been previously associated with AR; according to the study, males with homozygous SNP in IL9R are three times more likely to be affected than females. The effect could be assigned to either the presence of different IL9R splice variants influencing the IL9-binding ability or the involvement of IL9R in the early T cell development.[30]
Aschard et al. were able to show a significant effect of rs2069885 of IL9 in males on lung function and polysensitization.[31] On the other hand, Schuurhof et al. suggested that the IL9 genetic polymorphism (rs2069885) has an opposite effect on the risk of severe respiratory syncytial virus infection bronchiolitis in boys and girls[32] and Nouri-Aria et al. showed that IL9 is upregulated in the nasal mucosa during the pollen season and correlates with tissue infiltration by eosinophils.[33] Finally, Melén et al. showed sex-specific protective effects of IL9R haplotypes on childhood wheezing and sensitization. Their results suggest a role for IL9R in the pathophysiology of allergic diseases, which may be sex dependent.[34] Besides, Namkung et al. revealed an association between IL9R gene polymorphisms and atopic dermatitis. They showed that a genotype combination of IL9/IL9R genes significantly increased the risk of developing the AD phenotype.[8]
The IL17 families of cytokines have been linked to many diseases.[35,36] Elevation of plasma IL17 levels to >20 pg/mL has been considered an independent risk factor for severe asthma.[37] IL17A and IL17F can induce expression of several cytokines that have also been linked to asthma or asthma-related phenotypes.[38] One significant finding was the lack of an association between asthma and the examined SNPs of IL17F, including rs763780 (H161R). This SNP was associated with asthma in a Japanese population as one of its alleles was found to reduce the risk of asthma.[15] Ramsey et al. observed minimal contributions of 5 IL17F SNPs to asthma among white females.[39] These discrepancies could be due to differences in patient demographics, sample size, environmental factors, and genetic background, all of which can affect association studies.
The similar findings for IL17A and IL17F could well be due to either of the several factors:
• Coordinated chromatin modifications: Since both genes are located on the same chromosome (6p12), their promoters and the conserved noncoding sequence regions might have undergone coordinated chromatin modifications[40]
• Both genes act as homodimers or heterodimers and share similar biological functions, sequences, and expression patterns and many polymorphisms may influence IL17 expression.[41]
Therefore, the observed correlation between IL17F and IL17A genotype combination adds more validity to the results of this study. Recently, Wang et al. found an association among polymorphisms in the cytokine genes IL17A and IL17F and development of AR and severe asthma in Chinese cases.[17] IL17A rs2275913 has shown association with asthma phenotype in Chinese population.[41] However, another study on the association of IL17F rs763780 with asthma in European Americans, African Americans, and Saudi Arabian populations did not find any association.[39,42]
AR is genetically complex and may be influenced by several genetic and environmental components. IL9R belongs to the group of genes with a clear potential role in allergic diseases, and its modest associations with asthma have been reported.[26,43] IL9R is located on the pseudoautosomal region of X and Y chromosomes. Therefore, it seems to be biologically plausible to infer that the variants affect males (XY) and females (XX) differently. As the IL9 signaling pathway is involved in both airway and immunological processes, a possible role is suggested for the IL9R in the pathophysiology of AR. Further studies are warranted to further clarify the issue.
This study has a limitation on the number of participants due to lack of eligible patients in the study. In addition, it was not possible to apply the functional experiments because of impossibility of conducting these methods in our university.
Conclusion
Our data identify an association between the rs731476 SNP in the IL9R gene and the AR phenotype in the Iranian women while the combination of rs731476 T-/rs2069885 G in the IL9R/IL9 encoding genes led to a significantly higher risk for developing AR. IgE levels were not different between patients and controls. There are several limitations in our study including the sample size and lack of males group. The latter hampered from comparison of the results between the males within both groups.
As discussed above, determination of patient’s allelic state regarding the mentioned SNPs in these genes can improve management of patients’ care and quality of life. We recommend further studies with larger sample size in various ethnic groups on more candidate genes and functional studies. We also suggest Genome-wide Association Studies through next generation sequencing method.
Acknowledgements
We appreciate all patients for their great contribution to the study. This research has been supported by the Shahrekord University of Medical Sciences.
Financial support and sponsorship
The study was part a student project. There was no financial support.
Conflicts of interest
There are no conflicts of interest.
References
- Bousquet J, Khaltaev N, Cruz AA, Denburg J, Fokkens WJ, Togias A, et al. Allergic rhinitis and its impact on asthma (ARIA) 2008 update (in collaboration with the World Health Organization, GA(2) LEN and AllerGen). Allergy 2008;63 Suppl 86:8-160.
- Settipane RA, Charnock DR. Epidemiology of rhinitis: Allergic and nonallergic. Clin Allergy Immunol 2007;19:23-34.
- Feijen M, Gerritsen J, Postma DS. Genetics of allergic disease. Br Med Bull 2000;56:894-907.
- Dávila I, Mullol J, Ferrer M, Bartra J, del Cuvillo A, Montoro J, et al. Genetic aspects of allergic rhinitis. J Investig AllergolClin Immunol 2009;19 Suppl 1:25-31.
- Vermeesch JR, Petit P, Kermouni A, Renauld JC, Van Den Berghe H, Marynen P. The IL-9 receptor gene, located in the Xq/Yq pseudoautosomal region, has an autosomal origin, escapes X inactivation and is expressed from the Y. Hum Mol Genet 1997;6:1-8.
- Abdelilah S, Latifa K, Esra N, Cameron L, Bouchaib L, Nicolaides N, et al. Functional expression of IL-9 receptor by human neutrophils from asthmatic donors: Role in IL-8 release. J Immunol 2001;166:2768-74.
- Bauer JH, Liu KD, You Y, Lai SY, Goldsmith MA. Heteromerization of the gammac chain with the interleukin-9 receptor alpha subunit leads to STAT activation and prevention of apoptosis. J Biol Chem 1998;273:9255-60.
- Namkung JH, Lee JE, Kim E, Park GT, Yang HS, Jang HY, et al. An association between IL-9 and IL-9 receptor genepolymorphisms and atopic dermatitis in a Korean population. J Dermatol Sci 2011;62:16-21.
- De Smedt M, Verhasselt B, Kerre T, Vanhecke D, Naessens E, Leclercq G, et al. Signals from the IL-9 receptor are critical for the early stages of human intrathymic T cell development. J Immunol 2000;164:1761-7.
- Demoulin JB, Uyttenhove C, Van Roost E, DeLestré B, Donckers D, Van Snick J, et al. A single tyrosine of the interleukin-9 (IL-9) receptor is required for STAT activation, antiapoptotic activity, and growth regulation by IL-9. Mol Cell Biol 1996;16:4710-6.
- Little FF, Cruikshank WW, Center DM. Il-9 stimulates release of chemotactic factors from human bronchial epithelial cells. Am J Respir Cell Mol Biol 2001;25:347-52.
- Baraldo S, Faffe DS, Moore PE, Whitehead T, McKenna M, Silverman ES, et al. Interleukin-9 influences chemokine release in airway smooth muscle: Role of ERK. Am J Physiol Lung Cell Mol Physiol 2003;284:L1093-102.
- Paradowska-Gorycka A, Wojtecka-Lukasik E, Trefler J, Wojciechowska B, Lacki JK, Maslinski S. Association between IL-17F gene polymorphisms and susceptibility to and severity of rheumatoid arthritis (RA). Scand J Immunol 2010;72:134-41.
- Arisawa T, Tahara T, Shibata T, Nagasaka M, Nakamura M, Kamiya Y, et al. The influence of polymorphisms of interleukin-17A and interleukin-17F genes on the susceptibility to ulcerative colitis. J Clin Immunol 2008;28:44-9.
- Kawaguchi M, Takahashi D, Hizawa N, Suzuki S, Matsukura S, Kokubu F, et al. IL-17F sequence variant (His161Arg) is associated with protection against asthma and antagonizes wild-type IL-17F activity. J Allergy Clin Immunol 2006;117:795-801.
- Seiderer J, Elben I, Diegelmann J, Glas J, Stallhofer J, Tillack C, et al. Role of the novel Th17 cytokine IL-17F in inflammatorybowel disease (IBD): Upregulated colonic IL-17F expression in active Crohn’s disease and analysis of the IL17F p.His161Arg polymorphism in IBD. Inflamm Bowel Dis 2008;14:437-45.
- Wang M, Zhang Y, Han D, Zhang L. Association between polymorphisms in cytokine genes IL-17A and IL-17F and development of allergic rhinitis and comorbid asthma in Chinese subjects. Hum Immunol 2012;73:647-53.
- Schmidt-Weber CB, Akdis M, Akdis CA. TH17 cells in the big picture of immunology. J Allergy Clin Immunol 2007;120:247-54.
- Hellings PW, Kasran A, Liu Z, Vandekerckhove P, Wuyts A, Overbergh L, et al. Interleukin-17 orchestrates the granulocyte influx into airways after allergen inhalation in a mouse model of allergic asthma. Am J Respir Cell Mol Biol 2003;28:42-50.
- Prause O, Bozinovski S, Anderson GP, Lindén A. Increased matrix metalloproteinase-9 concentration and activity after stimulation with interleukin-17 in mouse airways. Thorax 2004;59:313-7.
- Hoshino H, Laan M, Sjöstrand M, Lötvall J, Skoogh BE, Linden A. Increased elastase and myeloperoxidase activity associated with neutrophil recruitment by IL-17 in airways in vivo. J Allergy Clin Immunol 2000;105(1 Pt 1):143-9.
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000;132:365-86.
- Doull IJ, Lawrence S, Watson M, Begishvili T, Beasley RW, Lampe F, et al. Allelic association of gene markers on chromosomes 5q and 11q with atopy and bronchial hyperresponsiveness. Am J Respir Crit Care Med 1996;153(4 Pt 1):1280-4.
- Rosenwasser LJ, Klemm DJ, Dresback JK, Inamura H, Mascali JJ, Klinnert M, et al. Promoter polymorphisms in the chromosome 5 gene cluster in asthma and atopy. Clin Exp Allergy 1995;25 Suppl 2:74-8.
- Nicolaides NC, Holroyd KJ, Ewart SL, Eleff SM, Kiser MB, Dragwa CR, et al. Interleukin 9: A candidate gene for asthma. Proc Natl Acad Sci U S A 1997;94:13175-80.
- Holroyd KJ, Martinati LC, Trabetti E, Scherpbier T, Eleff SM, Boner AL, et al. Asthma and bronchial hyperresponsiveness linked to the XY long arm pseudoautosomal region. Genomics 1998;52:233-5.
- Lu LF, Lind EF, Gondek DC, Bennett KA, Gleeson MW, Pino-Lagos K, et al. Mast cells are essential intermediaries in regulatory T-cell tolerance. Nature 2006;442:997-1002.
- Richard M, Grencis RK, Humphreys NE, Renauld JC, Van Snick J. Anti-IL-9 vaccination prevents worm expulsion and blood eosinophilia in Trichuris muris-infected mice. Proc Natl Acad Sci U S A 2000;97:767-72.
- Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, Sobel RA, et al. IL-4 inhibits TGF-beta-induced Foxp3+T cells and, together with TGF-beta, generates IL-9+IL-10+Foxp3(-) effector T cells. Nat Immunol 2008;9:1347-55.
- Burkhardt J, Petit-Teixeira E, Teixeira VH, Kirsten H, Garnier S, Ruehle S, et al. Association of the X-chromosomal genes TIMP1 and IL9R with rheumatoid arthritis. J Rheumatol 2009;36:2149-57.
- Aschard H, Bouzigon E, Corda E, Ulgen A, Dizier MH, Gormand F, et al. Sex-specific effect of IL9 polymorphisms on lung function and polysensitization. Genes Immun 2009;10:559-65.
- Schuurhof A, Bont L, Siezen CL, Hodemaekers H, van Houwelingen HC, Kimman TG, et al. Interleukin-9 polymorphism in infants with respiratory syncytial virus infection: An opposite effect in boys and girls. Pediatr Pulmonol 2010;45:608-13.
- Nouri-Aria KT, Pilette C, Jacobson MR, Watanabe H, Durham SR. IL-9 and c-Kit+mast cells in allergic rhinitis during seasonal allergen exposure: Effect of immunotherapy. J Allergy Clin Immunol 2005;116:73-9.
- Melén E, Gullstén H, Zucchelli M, Lindstedt A, Nyberg F, Wickman M, et al. Sex specific protective effects of interleukin-9 receptor haplotypes on childhood wheezing and sensitisation. J Med Genet 2004;41:e123.
- Chen Z, O’Shea JJ. Regulation of IL-17 production in human lymphocytes. Cytokine 2008;41:71-8.
- Starnes T, Broxmeyer HE, Robertson MJ, Hromas R. Cutting edge: IL-17D, a novel member of the IL-17 family, stimulates cytokine production and inhibits hemopoiesis. J Immunol 2002;169:642-6.
- Agache I, Ciobanu C, Agache C, Anghel M. Increased serum IL-17 is an independent risk factor for severe asthma. Respir Med 2010;104:1131-7.
- Wang YH, Liu YJ. The IL-17 cytokine family and their role in allergic inflammation. Curr Opin Immunol 2008;20:697-702.
- Ramsey CD, Lazarus R, Camargo CA Jr., Weiss ST, Celedón JC. Polymorphisms in the interleukin 17F gene (IL17F) and asthma. Genes Immun 2005;6:236-41.
- Pappu BP, Angkasekwinai P, Dong C. Regulatory mechanisms of helper T cell differentiation: New lessons learned from interleukin 17 family cytokines. Pharmacol Ther 2008;117:374-84.
- Chen J, Deng Y, Zhao J, Luo Z, Peng W, Yang J, et al. The polymorphism of IL-17 G-152A was associated with childhood asthma and bacterial colonization of the hypopharynx in bronchiolitis. J Clin Immunol 2010;30:539-45.
- Bazzi MD, Sultan MA, Al Tassan N, Alanazi M, Al-Amri A, Al-Hajjaj MS, et al. Interleukin 17A and F and asthma in Saudi Arabia: Gene polymorphisms and protein levels. J Investig Allergol Clin Immunol 2011;21:551-5.
- Kauppi P, Laitinen T, Ollikainen V, Mannila H, Laitinen LA, Kere J. The IL9R region contribution in asthma is supported by genetic association in an isolated population. Eur J Hum Genet 2000;8:788-92.




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.