In Vitro Evaluation Performance of Recently Developed Rapid Diagnostic Test for Detecting Carbapenemases: The Multiplex Lateral Flow Assay
Received: 16-Jan-2023, Manuscript No. AMHSR-23-108954; Editor assigned: 18-Jan-2023, Pre QC No. AMHSR-23-108954 (PQ); Reviewed: 01-Feb-2023 QC No. AMHSR-23-108954; Revised: 16-Jan-2025, Manuscript No. AMHSR-23-108954 (R); Published: 23-Jan-2025
Citation: Tarai B, et al. In Vitro Evaluation Performance of Recently Developed Rapid Diagnostic Test for Detecting Carbapenemases: The Multiplex Lateral Flow Assay. Ann Med Health Sci Res. 2025;15:1-4
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Carbapenem Resistant Enterobacteriaceae (CRE) and Carbapenemase- Producing Organisms (CPOs) pose a serious threat to public health. Due to their vast range of antibiotic resistance, which excludes carbapenems and affects the majority of antimicrobial agent classes, leaving relatively few alternatives to treat people who are infected. In this study, we compared the performance of the RESIST-5 O.K.N.V.I. and RESIST ACINETO test to Vitek-2 in detecting the carbapenemases among a large number of clinical isolates in India. Performance of ICT Assay in Comparison to Vitek, The IC test indings revealed 94.44% concordance and 2% discordant with the Vitek results with 94.44% sensitivity and 100 %speci icity. The RESIST-5 O.K.N.V.I. and RESIST ACINETO assay is quite effective at detecting carbapenemase genes. These assays are also a quick, simple, and effective technique for use in clinical microbiology laboratories.
Keywords
Antimicrobial agent; Carbapenem Resistant Enterobacteriaceae (CRE); Carbapenemase genes; Antibiotic resistance; Microbiology
Introduction
Specifically, Carbapenem-Resistant Enterobacteriaceae (CRE) and Carbapenemase-Producing Organisms (CPOs) pose a serious threat to public health. Due to their vast range of antibiotic resistance, which excludes carbapenems and affects the majority of antimicrobial agent classes, leaving relatively few alternatives to treat people who are infected. Along with CREs, CPOs also contain Nonfermenting Gram-Negative Bacilli (NFGNB), such as Pseudomonas aeruginosa and Acinetobacter baumannii, which show resistance to carbapenems in addition to beta-lactam and other antibiotic families. Worldwide nosocomial outbreaks and endemic conditions have been caused by the rapid spread of CPOs and the genes encoding these resistances [1]. Notably, genes for carbapenemases, which are thought to be a key factor in carbapenem resistance, are primarily found on mobile genetic elements such as transposons, plasmids, and genomic islands. Therefore, these genes are commonly transferred horizontally between bacterial species. It is difficult to quickly and accurately detect and identify carbapenemases in order to stop further spread and treat infected patients with suitable antimicrobials in the clinical setting remain a challenge [2].
Two of the most common and rapidly spreading carbapenemases worldwide are NDM and KPC. However, class D OXA48-type carbapenemases are the most difficult resistance mechanisms for clinical laboratories to identify. In addition to being found in Enterobacteriaceae, VIM is very common in non-fermenting bacteria. The Metallo-Lactamases (MBLs) of Ambler class B variant-NDM and OXA48-likevariants of Ambler class D, and the KPC variants of Ambler class A are the common carbapenemases that are most frequently found in Enterobacterales and non-fermenting bacterial isolates in India. Numerous diagnostic strategies, including culture-based approaches that use resistant phenotypes and molecular biology-based approaches that use gene amplification, have been extensively used in clinical microbiology laboratories to identify and characterise the varied kinds of carbapenemases [3-5]. However, the molecular method requires expensive equipment and high levels of knowledge, and culture-based phenotypic methods are laborintensive and time-consuming.
There is a need for the development of new rapid diagnostic tools to monitor antibiotic resistance patterns as one of the main core tasks [6].
The TRURAPID® O.K.N.V.I. RESIST-5 Test (manufactured by Kilpest India Ltd., marketed by 3B BlackBio Biotech India Ltd.) with membrane technology of colloidal gold nanoparticles was introduced to identify five targeted carbapenemase genes in a single test without the need for specialised equipment within 15 minutes. Another test, the TRURAPID® RESIST ACINETO Test (manufactured by Kilpest India Ltd., marketed by 3B BlackBio Biotech India Ltd.), which aims to quickly identify the OXA-23, OXA-40/58, and NDM carbapenemases (and variants of the OXA-23, 40, and 58 groups), guarantees efficient patient care and prevents the spread of carbapenemases Acinetobacter spp. carriers, particularly in hospitals. Recently, multiplex immunochromatographic lateral flow assays for detecting and characterising carbapenemases were developed [7,8].
In this study, we compared the performance of the TRURAPID® O.K.N.V.I. RESIST-5 Test and TRURAPID® RESIST ACINETO Test to Vitek-2 in detecting the carbapenemases among a large number of clinical isolates in India.
Materials and Methods
Study design
A total of 72 non-duplicated clinical isolates of gramnegative bacilli were included. The performance of the TRURAPID® O.K.N.V.I. RESIST-5 Test kit was assessed using 36 carbapenem-sensitive and 36 carbapenem-resistant bacterial isolates [9].
Bacterial identification and sensitivity
MALDI MS using a Biomeruix MALDI MS, was used to identify bacteria and all the isolates were characterized by VITEK-MS. Further, antibiotic sensitivity was done by Vitek-2.
TRURAPID® O.K.N.V.I. RESIST-5 Test Assay
These tests are ready to use and are based on a membrane technology with colloidal gold nanoparticles [10]. The kit is aimed to detect and identify the carbapenemases from a bacterial colony isolate of Enterobacteriaceae or NFGNB growing on an agar plate. Each pouch contains: 2 lateral-flow cassettes for the identification of (i) OXA-48, KPC, NDM and (ii) VIM and IMP.
Identification of OXA-48, KPC and NDM
A nitrocellulose membrane is sensitised with: (1) a monoclonal antibody directed against OXA-48 carbapenemase and variants (except OXA-163-like enzymes)(“O” line) (2) a monoclonal antibody directed against KPC carbapenemase (“K” line) (3) a monoclonal antibody directed against NDM carbapenemase (“N” line) (4) a control capture reagent (upper “C” line). Four different colloidal gold nanoparticles conjugates are dried on a membrane: A conjugate directed against a second epitope of the OXA-48 carbapenemase, a conjugate directed against a second epitope of the KPC carbapenemase, a third conjugate specific to NDM carbapenemase and a control conjugate to validate the test conditions [11].
Identification of VIM and IMP: A nitrocellulose membrane is sensitised with: (1) a monoclonal antibody directed against VIM carbapenemase (“V” line), (2) a monoclonal antibody directed against IMP carbapenemase (“I” line) (3) a control capture reagent (upper “C” line). Three different colloidal gold nanoparticle conjugates are dried on a membrane: A conjugate directed against VIM carbapenemase, a conjugate directed against IMP carbapenemase and a control conjugate [12].
When the provided buffer containing the resuspended bacteria comes into contact with the membrane, the solubilised conjugates migrate with the sample by passive diffusion, while conjugates and sample material come into contact with the immobilised respective antibodies that are adsorbed onto the nitrocellulose strip. If the sample contains an OXA-48, KPC, NDM, VIM or IMP carbapenemase, the respective complexes made of the conjugates and either OXA-48, or KPC, or NDM or VIM or IMP will remain bound to their respective specific lines (OXA-48 : “O” line; KPC : “K” line; NDM : “N” line, VIM : “V” line, IMP : “I” line). The migration continues by passive diffusion and both conjugates and sample material come into contact with the (upper) line control reagent that binds a control conjugate (“C” line), thereby producing a red line. The result is visible within 15 minutes in the form of red lines on the strip [13].
Statistical analysis
Data were collected and analyzed using STATA/SE 14.0 statistical software (Stata Corp LLC, 4905 Lakeway Drive college station, Texas, USA). Categorical data were described using numbers and percentages. Our data were presented in the form of tables, and descriptive statistics were analyzed in percentages [14]. P value was calculated using the Chi square test to analyze statistical significances.
Results
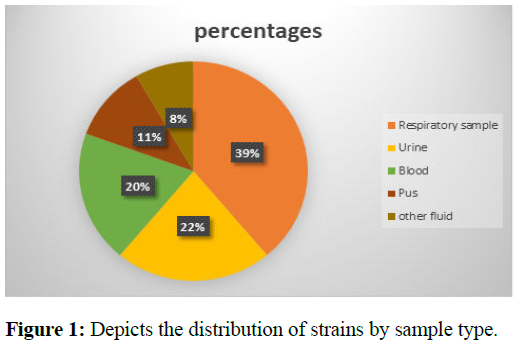
A total of 76 clinical isolates including both carbapenemsensitive and carbapenem-resistant strains were included in the study. Among the 76 clinical isolates, 36 were carbapenem-sensitive and 40 were carbapenem-resistant. All 40 carbapenem-resistant isolates were correctly identified by the TRURAPID® O.K.N.V.I. RESIST-5 Test assay compared to the conventional method. Out of 40 carbapenemaseproducing organisms, 30 were carbapenem-resistant Enterobacteriaceae and 16 were carbapenem-resistant Non- Fermenter Gram-Negative Bacilli (NFGNB). Figure 1 shows the distribution of strains according to the types of samples which were predominant by respiratory sample (39%) and urine (22%) samples.
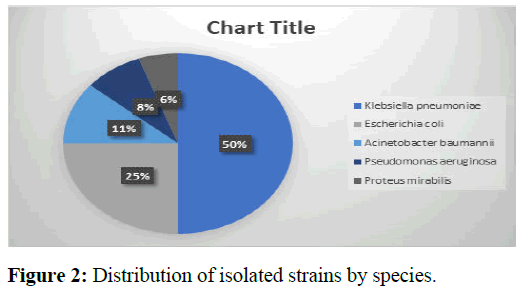
The species found were predominantly Klebsiella pneumoniae 50%, followed by Escherichia coli 25% and Acinetobacter baumannii 11% as shown in Figure 2 [15].
Performance of ICT Assay in Comparison to Vitek, The IC test findings revealed 94.44% concordance and 2%discordant with the Vitek results with 94.44% sensitivity and 100 % specificity.
NDM was detected in these strains most frequently in the ICT tests, or 23/40 (57.50%), followed by OXA-48 in 20/40 (50%), and 13/40 (32.50%) of cases when OXA-48 and NDM coexisted. Out of 10 Acinetobacter species, only one was positive for NDM by TRURAPID® O.K.N.V.I. RESIST-5 Test assay, further, all Acinetobacter species were tested by TRURAPID® RESIST ACINETO Test assay [16]. It has been observed that 9/10 Acinetobacter species were positive for OXA-23 and one Acinetobacter species was positive for both OXA-23 and OXA-40/58. In contrast, 2/40(5%) of the cases were negative, which were positive by Vitek (Table 1).
| Vitek + | Vitek - | Total | |
|---|---|---|---|
| ICT + | 38 | 0 | 38 |
| ICT - | 2 | 0 | 2 |
| Total | 40 | 0 | 40 |
Table 1: ICT assay performance as compared with Vitek.
Discussion
The findings of the IC test revealed that Vitek-2 and the detection of carbapenemases were 94.44% compatible. They were also successful in concurrently detecting two carbapenemases with 94.44% sensitivity and 100 % specificity and no cross-reactivity [17].
Our research revealed the formation of the NDM strain, the dominance of OXA-48, and instances of coexisting carbapenemases (OXA-48 and NDM). Our findings are consistent with those of Bianco et al., who discovered OXA-48 and NDM-like carbapenemases from 4-hour subcultures on blood culture samples with 100% sensitivity [18]. Additionally, they concur with the findings of the investigation by Han et al., in which OXA-48 had a detection sensitivity of 100% but NDM had a detection sensitivity of 97.1%. Additionally, studies using the RESIST-4 O.K.N.V. (OXA-48, KPC, NDM, and VIM), RESIST-3 O.K.N. (OXA-48, KPC, and NDM), OXA-163/-48 K-Se T, and OXA-48 K-SeT based on the same principle as the RESIST-5 O.O.K.N.V. have discovered a sensitivity of 100% for OXA-48 and a sensitivity for NDM ranging from 97.8% to 100%. We were unable to evaluate the effectiveness of the RESIST-5 O.O.K.N.V. for their detection in the absence of OXA-23 and OXA-40/58 producing Acinetobacter species, which were further detected by the TRURAPID® RESIST ACINETO test [19].
The IC RESIST-5 O.O.K.N.V test does not require substantial expertise, specialised labs, or high-tech equipment to perform as does the PCR and sequencing test. The test is quick to run (less than 5 minutes), reliable, affordable, and simple to use in all laboratories for the detection of carbapenemases. However, they fail to identify unusual carbapenemases like GES, IMI, and IMP because they only identify common carbapenemases.
For patients with infections brought on by CPE, the IC test may be a quick and accurate tool to expedite management, antibiotic administration, and infection control measures. In fact, the quick identification of carbapenemases enables rapid optimisation of empirical antibiotic therapy based on local epidemiology and the prediction of multidrug-resistant phenotypes [20].
In actuality, ceftazidime-avibactam is effective against the majority of Enterobacterales that produce KPC or OXA-48 type carbapenemase. Contrarily, metallo-beta lactamases (NDM, VIM, or IMP) are not inhibited by ceftazidimeavibactam; as a result, alternative treatment methods such as tigecycline, colistin, or other novel antimicrobial agents (such as aztreonam-avibactam) must be used.
Conclusion
According to our findings, the TRURAPID® O.K.N.V.I. RESIST-5 Test and TRURAPID® RESIST ACINETO Test assays are quite effective at detecting carbapenemase genes. These assays are also a quick, simple, and effective technique for use in clinical microbiology laboratories. In order to assist stop the spread of CPEs, the hospital should also regularly implement antibiotic susceptibility databases and national, regional, and local surveillance studies.
References
- Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L. Carbapenemase-producing organisms: A global scourge. Clin Infect Dis. 2018;66:1290-1297.
[Crossref] [Google Scholar] [PubMed]
- Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: The impact and evolution of a global menace. J Infect Dis. 2017;215:S28-S36.
[Crossref] [Google Scholar] [PubMed]
- Hong JS, Yoon EJ, Lee H, Jeong SH, Lee, K. Clonal dissemination of pseudomonas aeruginosa sequence type 235 isolates carrying blaIMP-6 and emergence of blaGES-24 and blaIMP-10 on novel genomic islands PAGI-15 and -16 in South Korea. Antimicrob Agents Chemother. 2016;60:7216-7223.
[Crossref] [Google Scholar] [PubMed]
- van Duin D, Doi Y. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 2017;8:460-469.
[Crossref] [Google Scholar] [PubMed]
- Maurer FP, Castelberg C, Quiblier C, Bloemberg GV, Hombach M. Evaluation of carbapenemase screening and confirmation tests with Enterobacteriaceae and development of a practical diagnostic algorithm. J Clin Microbiol. 2015;53:95-104.
[Crossref] [Google Scholar] [PubMed]
- Kim H, Sung JY, Yong D, Jeong SH, Song W, et al. Disk carbapenemase test for the rapid detection of KPC-, NDM-, and Other Metallo-β-Lactamase-Producing Gram-Negative Bacilli. Ann Lab Med. 2016;36:434-440.
[Crossref] [Google Scholar] [PubMed]
- Hong JS, Kim D, Yoon EJ, Lee H, Jeong SH. Performance evaluation of the PANA RealTyper™ CRE Kit for detecting carbapenemase genes in Gram-negative bacilli. J Glob Antimicrob Resist. 2019;18:100-103.
[Crossref] [Google Scholar] [PubMed]
- Bogaerts P, Berger AS, Evrard S, Huang TD. Comparison of two multiplex immunochromatographic assays for the rapid detection of major carbapenemases in Enterobacterales. J Antimicrob Chemother. 2020;75:1491-1494.
[Crossref] [Google Scholar] [PubMed]
- Boutal H, Vogel A, Bernabeu S, Devilliers K, Creton E, et al. A multiplex lateral flow immunoassay for the rapid identification of NDM-, KPC-, IMP and VIM-type and OXA-48-like carbapenemase-producing Enterobacteriaceae. J Antimicrob Chemother. 2018;73:909-915.
[Crossref] [Google Scholar] [PubMed]
- Bianco G, Boattini M, van Asten S, Iannaccone M, Zanotto E, et al. eRESIST-5 O.O.K.N.V. and NG-Test Carba 5 assays for the rapid detection of carbapenemase-producing Enterobacterales from positive blood cultures: A comparative study. J Hosp Infect. 2020;105:162-166.
[Crossref] [Google Scholar] [PubMed]
- Glupczynski Y, Jousset A, Evrard S, Bonnin RA, Huang TD, Dortet L, et al. Prospective evaluation of the OKN K-SeT assay, a new multiplex immunochromatographic test for the rapid detection of OXA-48-like, KPC and NDM carbapenemases. J Antimicrob Chemother. 2017;72:1955-1960.
[Crossref] [Google Scholar] [PubMed]
- Wareham DW, Abdul Momin MH. Rapid detection of carbapenemases in Enterobacteriaceae: evaluation of the Resist-3 OKN (OXA-48, KPC, NDM) lateral flow multiplexed assay. J Clin Microbiol. 2017; 55:1223-1225.
[Crossref] [Google Scholar] [PubMed]
- Meunier D, Vickers A, Pike R, Hill RL, Woodford N, Hopkins KL. Evaluation of the K-SeT RESIST immunochromatographic assay for the rapid detection of KPC and OXA-48-like carbapenemases. J Antimicrob Chemother. 2016;71:2357-2359.
[Crossref] [Google Scholar] [PubMed]
- Wareham DW, Shah R, Betts JW, Phee LM, Momin MH. Evaluation of an immunochromatographic lateral flow assay (OXA-48 K-SeT) for rapid detection of OXA-48-like carbapenemases in Enterobacteriaceae. J Clin Microbiol. 2016;54: 471-473.
[Crossref] [Google Scholar] [PubMed]
- Fernández J, Fleites A, Rodcio MR, Vazquez F. Evaluation of OXA-48 K-Se T: an immunochromatographic assay for rapid detection of OXA-48-producing Enterobacteriaceae. Diagn Microbiol Infect Dis. 2016;85:12-15.
[Crossref] [Google Scholar] [PubMed]
- Dortet L, Jousset A, Sainte-Rose V, Cuzon G, Naas T. Prospective evaluation of the OXA-48 K-SeT assay, an immunochromatographic test for the rapid detection of OXA-48-type carbapenemases. J Antimicrob Chemother. 2016;71:1834-1840.
[Crossref] [Google Scholar] [PubMed]
- Glupczynski Y, Evrard S, Ote I, Mertens P, Huang TD, Leclipteux T, Bogaerts P. Evaluation of two new commercial immunochromatographic assays for the rapid detection of OXA-48 and KPC carbapenemases from cultured bacteria. J Antimicrob Chemother. 2016;71:1217-1222.
[Crossref] [Google Scholar] [PubMed]
- Song W, Park MJ, Jeong S, Shin DH, Kim JS, Kim HS, et al. Rapid identification of OXA-48-like, KPC, NDM, and VIM carbapenemase-producing Enterobacteriaceae from culture: Evaluation of the RESIST-4 OKNV multiplex lateral flow assay. Ann Lab Med. 2020;40:259-263.
[Crossref] [Google Scholar] [PubMed]
- Falcone M, Paterson D. Spotlight on ceftazidime/avibactam: A new option for MDR Gram-negative infections. J Antimicrob Chemother. 2016;71:2713-22.
[Crossref] [Google Scholar] [PubMed]
- Bassetti MA, Giacobbe DR, Giamarellou H, Viscoli C, Daikos GL, Dimopoulos G, et al. Management of KPC-producing Klebsiella pneumoniae infections. Clin Microbiol Infect. 2018;24:133-144.
[Crossref] [Google Scholar] [PubMed]




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.