Increased Virus Replication and Cytotoxicity of Non‑pathogenic Simian Human Immuno Deficiency Viruses‑NM‑3rN After Serial Passage in a Monkey‑Derived Cell Line
- *Corresponding Author:
- Dr. Theophilus Kwofie
Department of Clinical Microbiology, School of Medical Sciences, College Health Science, Kwame Nkrumah University of Science and Technology, Kumasi, Ghana
E-mail: tbenkwofie@yahoo.com
Abstract
Background: Infection and disease induction of variants of HIV type 1 (HIV‑1) in vivo, especially their persistence, replication and rate of disease progression, have been found to depend on phenotypic characteristics. However, the mechanism (s) underlying these diverse phenotypic characteristics remain poorly understood. Aim: It was aimed at determining whether a SHIV that had been adapted to a monkey‑derived cell line could be used to explain the mechanism that underlies adaptive evolution of a virus to its host cell environment. Materials and Methods: Standard procedures in virology such as cell culturing, FACS analysis and ELISA were employed to measure virus replication and growth kinetics, cell viability, reverse transcriptase (RT) activity assay and CD4 cells down‑regulation. Results: After about 20 passages, LT efficiently adapted to the monkey‑derived cell line and replicated much better than the parent virus. LT accumulated a number of mutations in its entire genome with a majority of them being monkey cell‑specific. Conclusion: Thus we think we have obtained a virus that may enable studies to determine which of these mutations are specifically related to in vitro viral replication and which are specifically related to cytotoxicity so as to explain the mechanism associated with viral cytotoxicity and host cell specificity.
Keywords
Cell culture, Evolution, HIV/SIV, Monkey, Replication
Introduction
Infection and disease induction of variants of HIV type 1 (HIV-1) in vivo, especially their persistence, replication and rate of disease progression, have been found to depend on phenotypic characteristics, such as host cell tropism, co-receptor usage, replication rate, cytopathicity and syncytium induction.[1-4] However, the mechanism (s) underlying these diverse phenotypic characteristics remain poorly understood partly due to the unavailability of suitable animal models for HIV.[5-10] Therefore, chimeric simian/ human immunodeficiency viruses (SHIVs) that express HIV-1 envelopes on an simian immunodeficiency virus (SIV) backbone have been developed.[11-14] These SHIVs display functional diversity, especially in their ability to infect and replicate in different cell types as well as their ability to induce cytopathicity.[11-14] Those SHIVs that display pathogenic properties are able to infect macaque monkeys though the disease they course do not necessarily mimic HIV infection in humans.[15-18] One such SHIV, SHIV-NM-3rN, is a chimeric virus from SIVmac239 and HIV-1NL432 that carries the env, tat, rev, vpr and vpu from the HIV-1 and the other regions from the SIVmac.[19,20] SHIV-NM-3rN is considered non-pathogenic virus, as infection in non-human primates does not lead to disease.[20] Also, NM-3rN does not replicate to high titers in human or macaque PBMCs or in monkey-derived cell lines such as HSC-F cells, although it does readily infect and replicate in human-derived cell lines such as M8166 cells.[20] NM-3rN maintained on M8166 cells for more than 20 passages is able to minimally replicate in HSC-F cells. We refer to the M8166-maintained virus as NM-3rN (M). Further continuous passage of SHIV-NM-3rN (M) in HSC-F cells resulted in virus progeny with increased replication rates and cytotoxicity. This long-term-passaged, monkey cell-adapted SHIV-NM-3rN was named NM-3rN (LT), or LT. The purpose of the present study was to identify the mechanism (s) underlying the change of NM-3rN (M) from a slow replicating virus on HSC-F cells to a variant NM-3rN (LT) with high replication rates on these same cells. We present evidence that critical mutations in LT viral genome may be responsible for the increased replication in the monkey-derived cell lines.
Materials and Methods
Viruses
The construction and the biological and virological properties of SHIV-NM-3rN were described previously.[20] SHIV-NM-3rN is a non-pathogenic molecularly cloned chimeric virus constructed from SIVmac 239 and HIV-1 NL432, which carries the env, tat, rev, vpr and vpu from HIV-1 and the other regions from SIVmac. SHIV-NM-3rN acquired a few mutations in its HIV-1 component, thus making this component less homologous to the parental HIV-1 NL432. The entire sequence of SHIV-NM-3rN has been submitted to GenBank (accession number AB177846). While this virus readily infects and replicates in M8166 cells (a human-derived cell line), it cannot replicate in HSC-F cells (a monkey derived cell line).
Cells
This research was purely an in vitro cell culture experiment not involving any human or animal use and therefore did not require any formal ethical clearance; however, appropriate disciplinary ethical guidelines of our institution was followed. PBMCs were prepared from 10 ml of blood from an HIV sero-negative human and a SIV sero-negative monkey. PBMCs were obtained from the blood samples by density gradient centrifugation. The PBMCs were stimulated with Concanavalin A (ConA) for 24 hours and then maintained in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (GIBCO-BRL), 20 mM L-glutamine, 400 units/ml IL-2, 10 mM sodium pyruvate, 5 × 10 5 M 2-ME (Mercaptoethanol) and antibiotics. M8166 and HSC-F cells were also maintained in the same medium without IL-2 and sodium pyruvate. HSC-F cells were established from CD4+ T cells derived from the spleen of the fetus of a cynomolgus monkey.[21] The CD4+ T cells, which express some markers of activated or immature T cell phenotypes such as CD4 and CD8, were immortalized by infection with herpes virus saimiri subtype C. M8166 cells are clones derived through limiting dilution of C8166 cells.[22]
Growth kinetics and reverse transcriptase activity assay
Replication kinetics of the viruses were examined on human and monkey PBMCs, and M8166 and HSC-F cells by inoculating duplicate cultures of 5 × 105 cells with 100 TCID50 of each virus in a 96-well plate. After inoculation, the culture supernatant was collected every 3 days and assessed for viral replication by judging the RT activity as previously described.[23]
Down-regulation of CD4 cells
The ability of the viruses to down-regulate CD4 cells in vitro was studied in HSC-F cells. One hundred TCID50 of each virus was infected to duplicate cultures of 5 × 105 HSC-F cells/well in a 12-well micro-plate. Five hundred microliters of the virus infected-cell suspension was transferred to a new well and an equivalent amount of fresh cells was added every three days. The remaining virus-infected cells were used for FACS analysis as described elsewhere.[13]
Count of viable cells
The effect of the viruses on cell viability through suppressing of cell division and/or cell killing was examined in monkey PBMC and HSC-F cells. One hundred TCID50 of each virus was infected to the cells in a 96-well micro-plate. Plates were incubated at 37°C for 3 days after which 10 μl of WST-8 reagent (Dojindo Laboratories, Kumamoto, Japan) was added to each well and incubated for an additional 3 hours. The percentage of viable cells in the plate, which corresponds to the intensity of the color of the medium on the cells, was determined with an ELISA plate reader.
Relationship between cellular infectivity and CD4 down-modulation
The relationship between the frequency of infected cells and the percentage of the simultaneous reduction of the CD4 surface marker was determined as follows. One hundred TCID50 of each virus was infected to duplicate cultures of 5 × 105 HSC-F cells/well in a 12-well micro-plate. Five hundred microliters of this virus infected-cell suspension was transferred to a new well and an equivalent amount of fresh cells was added every three days. For a FACS analysis, 5 × 105 cells were pelleted in a 1.5-ml Eppendorf tube, washed once with trypsin/EDTA solution to remove unbound virus from the cell surface, washed two times in wash buffer (PBS containing 0.5% bovine serum albumin (BSA)), suspended in 50 μl of the wash buffer, labelled with 20 μl of reactive SHIV-infected monkey anti-serum, incubated for 30 min on ice, washed once, stained with 10 μl each of 10 × diluted monoclonal antibodies CD4-PE, CD8-perCP and FITC-labelled anti-sera (IgA, IgG, and IgM) (Becton Dickinson, Erembodegem, Belgium), incubated for 30 minutes. In the dark, washed two times in the same wash buffer and re-suspended in PBS containing 1% BSA and 10% paraformaldehyde. The percentage of CD4 and viral protein cells was determined using FACScan (Becton Dickinson, Erembodegem, Belgium).
Sequence analysis
Direct sequencing was performed on PCR products generated from the whole genome of the virus using an automated BigDye system (ABI PRISM 310 genetic analyzer; PerkinElmer, Emeryville, CA). Database searches were carried out manually with the aid of Genetyx-Mac software.
Results
After 20 or more passages in HSC-F, NM-3rN became a more efficient virus, at infecting and replicating in HSC-F cells The new virus, NM-3rN (LT)(LT) exhibited other distinct biological properties in its replication kinetics, cytopathicity, and syncytium-induction.
Replication kinetics
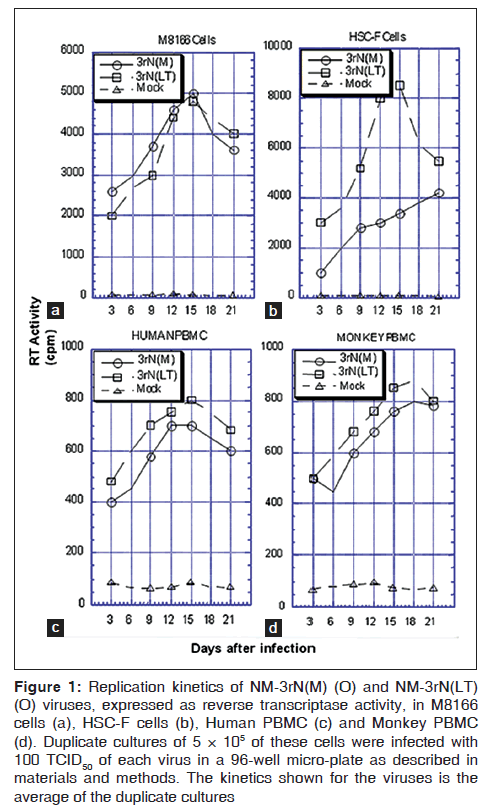
Replication ability was indirectly determined by measuring reverse transcriptase (RT) activity. The replication kinetics of LT and NM-3rN (M) in M8166 cell were similar [Figure 1a].
Figure 1: Replication kinetics of NM-3rN(M) (O) and NM-3rN(LT) (O) viruses, expressed as reverse transcriptase activity, in M8166 cells (a), HSC-F cells (b), Human PBMC (c) and Monkey PBMC (d). Duplicate cultures of 5 × 105 of these cells were infected with 100 TCID50 of each virus in a 96-well micro-plate as described in materials and methods. The kinetics shown for the viruses is the average of the duplicate cultures
However, in HSC-F cells, peak RT activity of LT (>8000 cpm on day 15) was much greater than that of NM-3rN (M) (3000 cpm on day 15) [Figure 1b]. LT also replicated better in human and simian PBMCs although to a lesser extent [Figures 1c and d]. However, no change was observed in the co-receptor usage of LT. LT, like NM-3rN (M), replicated only in CXCR4-expressing cells (results not shown).
Cytopathicity
LT demonstrated more cytopathicity than NM-3rN (M). For example LT decreased the CD4 positive cells on HSC-F cells to a greater extent (78% at day 3 and 82% at day 12) than NM-3rN (M) (1% at day 3 and 47% at day 12) [Figure 2]. Furthermore, 3 days after infection, there was a reduction in the number of viable cells in the LT infected monkey PBMC and HSC-F cells by as much as 20% and 50% respectively. In contrast the reduction in the number of viable cells in the NM-3rN (M)-infected cultures was 8% and 17% respectively [Figure 3]. In addition LT was much more effective than NM-3rN (M) in inducing syncytia in HSC-F and M8166 cells (results not shown). Thus, LT consistently displayed a more pathogenic phenotype than NM-3rN (M).
Figure 2: This is a figure showing the down-regulation of the CD4+ cell population by the viruses {NM-3rN(M) and NM-3rN(LT)} in HSC-F cells. The CD4+ cell populations at the indicated time points 3, 6, 9 and 12 days post infection were monitored by FACS analysis as explained in the materials and methods
Relationship between cellular infectivity and CD4 down-modulation
LT rapidly infected and replicated in HSC-F cells and this correlated with faster reduction of surface CD4 markers in these cells than did NM-3rN (M). For example, 3 days post infection, LT had infected 71% of the cells with an associated reduction in cell surface CD4 markers by about 16%, while NM-3rN (M) had infected only 35% of the cells with 1% reduction in cell surface CD4. At day 9 post infection, LT had infected almost all the cells with an 80% reduction in cell surface CD4 while NM-3rN (M) infected about 78% of the cells with an associated reduction in cell surface CD4 by only 13% [Figures 4a and b].
Figure 4: These figures show the relationship between the infectivity of the viruses NM-3rN (M) and (LT) and their corresponding down-regulation of the CD4+ cells monitored in HSC-F cells as described in the materials and methods. (a) the percentage of the cells that were infected at the indicated time points while (b) the percentage of the CD4+ cell remaining in the culture at the same time points
Sequence analysis
The sequence of the LT genome is compared with the sequences of the plasmid SHIVNM- 3rN and NM-3rN (M) before its adaptation to HSC-F cells in Table 1. Before NM- 3rN (M) was introduced to HSC-F cells, it had acquired 12 nucleotide changes in its genome [shown in color in column 3 of Table 1], possibly through long-term passage in M8166 cells. However, after the virus adapted to HSC-F cells, three of the mutations (brown) reverted back to the original sequence. Of the remaining 9 mutations, 5 (green) were retained and 4 (blue) further mutated. LT also acquired 21 other mutations throughout its genome [yellow nucleotides in Table 1]. Some of these mutations were silent while others resulted in amino acids changes. The percentage of the amino acids in the original plasmid NM-3rN that were changed ranged from 0.2% to 1.4% in the pol, tat and nef genes and from 2.0% to 3.2% in the gag, vpr, rev and env genes. LT had a 12-amino acid frame shift deletion from its vpr gene to tat gene and another 5-amino acid deletion in the gp120 region of the env gene. Among the 5 NM-3rN (M) mutations that were retained in LT, one was at position 8560 of the env gene, b which resulted in a stop codon at that position. LT had another stop codon at position 6222 of its vpu gene.
| Location(nt# in pNM3rN) | pNM-3rNA | NM-3rN M) B | NM-3rN (LT) C | Gene | Amino Acid | |
|---|---|---|---|---|---|---|
| 181 | T | T | G | U5 | ||
| 190 | T | T | C | U5 | ||
| 312 | T | C | C | none coding | ||
| 543 | T | T/C | T/C | gag p17 | V/A | |
| 659 | G | G/A | G | gag p17 | D/N | |
| 663 | G | G/A | A | gag p17 | R/K | |
| 677 | G | G/A | A | gag p17 | E/K | |
| 1516 | T | T | C | gag p24 | A/A | silent |
| 2245 | A | A | G | pol pr | R/G | |
| 2948 | C | C | T | pol RT | S/L | |
| 5858 | G | G | C | vpr | G/G | silent |
| 5859-5895 | NODEL | NODEL | DEL | vpr | 12aa del frameshift to tat | |
| 6053 | A | A | G | tat | K/R | |
| rev | R/G | |||||
| 6222 | T | T | A | vpu | Y/stop | |
| 6654 | T | T/C | C | env gp120 | V/A | |
| 6678 | G | G/A | G | env gp120 | S/N | |
| 6745 | G | G | A | env gp120 | E/E | silent |
| 6805 | G | G | A | env gp120 | Q/Q | silent |
| 7401 | T | T | C | env gp120 | I/T | |
| 7475-7489 | NODEL | NODEL | NODEL/DEL | env gp120 | 5aa del | |
| 7574 | G | G | A | env gp120 | E/K | |
| 7682 | T | T | C | env gp120 | S/P | |
| 7929 | A | G | G | env gp41 | D/G | |
| 8159 | A | A | A/G | env gp41 | N/D | |
| 8200 | T | G | G | env gp41 | N/K | |
| 8412 | T | T | T/C | env gp41 | V/A | |
| 8425 | T | T | C | env gp41 | Y/Y | silent |
| 8560 | G | A | A | env gp41 | W/stop | |
| 8720 | G | G | A | env gp41 | A/T | |
| 8936 | G | G | A | nef | G/G | silent |
| 8937 | G | G | A | nef | E/K | |
| 9486 | G | G/A | A | nef | E/K | |
| 9534 | G | G/A | G | nef | A/T |
Table 1: Sequence comparison of the whole genomes of pSHIV-NM-3rN, NM-3nN (M) AND NM-3rN (LT)
Discussion
This study was undertaken to identify the mechanism (s) that may be underlying the change of a slow replicating SHIV, SHIV-NM-3rN (M), on a monkey-derived cell line to a variant, NM-3rN (LT) which could replicate better on these same cells. It was to determine whether a SHIV that had been adapted to a monkey-derived cell line could be used to explain the mechanism that underlies adaptive evolution of a virus to its host cell environment. We believe that the identification of such mechanism (s) could be used to explain the pathogenicity of these SHIVs and the mechanism that underlies evolution of adaptation.
After about 20 passages, LT efficiently adapted to the monkey derived cell line and replicated better than the parent virus, NM-3rN (M). Some of the mutations that took place in the LT genome, as a result of its prolonged passage in HSC-F cells, might have resulted in a favourable interaction between the genes of LT and HSC-F cells. This would have enabled LT to take significantly less time than NM-3rN (M) to infect and spread to the majority of the cells. The reduction in HSC-F surface CD4 markers was independent of infection and that CD4 down-modulation was probably virus specific rather than due to the infection per se as evidenced by the uneven infection of the viruses in HSC-F cells [day 3 of Figure 4a]. This could be due to LT replicating better than NM-3rN (M) because at day 6, when NM-3rN (M) had infected about 70% of the cells, it did not down-modulate the cell surface CD4 makers. Even at day 9, when LT had infected almost all the cells and reduced their surface CD4 markers by about 80%, NM-3rN (M) had infected about 80% of the cells and reduced their CD4 surface markers by just about 13% [Figures 4a and 4b]. This suggests that LT had adapted to the monkey-derived cells, and it did so by acquiring much better cellular activation signals that enabled it to initiate and maintain replication in these cells. Although NM-3rN is a cloned virus continuous passaging in HSC-F cells may have caused mutations resulting in a mixed viral population. These viral variants may have subsequently been clonally selected and amplified based on their fitness resulting in the dominance and survival of one clone, in this case LT. Together, these results suggest that the enhanced in vitro replication of LT may have been due to both adaptation and selection. The 12 mutations that were present in NM-3rN (M) before its introduction into HSC-F cells are of interest because they likely were a result of NM-3rN’s long-term passage in M8166 cells and that only the long-term M8166 cell-passage virus could infect HSC-F cells, albeit minimally. Therefore, mutations acquired in M8166 contributed to NM-3rN (M)’s ability to infect HSC-F cells and are likely essential for this virus to grow in established cell lines especially as NM-3rN without intermediate M8166 passages has been found not able to directly infect HSC-F cells.[11] The enhanced replication properties of LT in HSC-F cells are most likely due to the 21 mutations it acquired in HSC-F cells implying LT had accumulated a number of mutations throughout its genome most of which were monkey-specific. Our results suggest that these mutations are responsible for the increased replication of LT.
LT therefore may be a valuable and interesting virus because it may enable studies to determine which of these mutations are specifically related to viral replication and which are specifically related to cytotoxicity. We do, however, recognized that in vivo conditions are not the same as in vitro and therefore an in vitro-derived virus may not necessary exhibit the same properties when introduce to in vivo studies, none-the-less, we think further studies and manipulation of LT may enabled it to be used to explain the functional diversity of HIVs and to understand how a virus adapts to a new host. Thus, we think we have obtained a virus that can be used to identify markers that are associated with viral cytotoxicity and host cell specificity.
Source of Support: Nil.
Conflict of Interest: None declared.
References
- Rudensey LM, Papenhausen MD, Overbaugh J. Replication and persistence of simian immunodeficiency virus variants after passage in macaque lymphocytes and established human cell lines. J Virol 1993;67:1727-33.
- Fenyo EM, Albert J, Asjo B. Replicative capacity, cytopathic effect and cell tropism of HIV. AIDS 1989;3:5-12.
- Greenier JL, Miller CJ, Lu D, Dailey PJ, Lu FX, Kunstman KJ, et al. Route of simian immunodeficiency virus inoculation determines the complexity but not the identity of viral variant populations that infect rhesus macaques. J Virol 2001;75:3753-65.
- Kestler HWD, Ringler DJ, Mori K, Panicali DL, Sehgal PK, Daniel MD, et al. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 1991;65:651-62.
- Baba TW, Jeong YS, Pennick D, Bronson R, Greene MF, Ruprecht RM. Pathogenicity of live attenuated SIV after mucosal infection of neonatal macaques. Science 1995;267:1820-5.
- Dunn CS, Hurtrel B, Beyar C, Gloeckler L, Ledger TN, Moog C, et al. Protection of SIVmac-infected macaque monkeys against super infection by a simian immunodeficiency virus expressing envelope glycoproteins of HIV type 1. AIDS Res Hum Retroviruses 1997;13:913-22.
- Giavedoni LD, Planelles V, Haigwood NL, Ahmad S, Kluge JD, Marthas ML, et al. Immune response of rhesus macaques to recombinant simian immunodeficiency virus gp 130 does not protect from challenge infection. J Virol 1993;67:577-83.
- Asjo B, Morfeldt-Manson L, Albert J, Biberfeld G, Karlsson A, Lidman K, et al. Replicative capacity of human immunodeficiency virus from patients with varying severity of HIV infection. Lancet 1986;ix: 660-2.
- Cloyd, MW, Moore BE. Spectrum of biological properties of human immunodeficiency virus (HIV-1) isolates. Virology 1990;174:103-16.
- Dahl K, Martin K, Miller G. Differences among human immunodeficiency virus strains in their capacities to induce cytolysis or persistent infection of a lympho-blastoid cell line immortalized by Epstein-Barr virus. J Virol 1987;61:1602-8.
- Kuwata T, Shioda T, Igarashi T, Ido E, Ibuki K, Enose Y, et al. Chimeric viruses between SIVmac and various HIV-1 isolates having biological properties that are similar to those of the parental HIV-1. AIDS 1996;10:1331-7.
- Reimann KA, Li JT, Veazey R, Halloran M, Park I-W, Karlsson GB, et al. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol 1996;70:6922-8.
- Ui M, Kuwata T, Igarashi T, Ibuki K, Kozyrez IL, Enose Y, et al. Protection of macaques against a SHIV with homologous HIV-1 env and a Pathogenic SHIV 89.6P with a Heterologous env by vaccination with multiple gene-deleted SHIVs. Virology 1999;265:252-63.
- Li J, Lord CI, Haseltine W, Letvin NL, Sodroski J. Infection of cynomolgus monkeys with a chimeric HIV-1/SIVmac virus that expresses the HIV-1 envelope glycoproteins. J Acquir Immune Defic Syndr 1992;5:639-46.
- Bangham CR, Philips RE. What is required of an HIV vaccine. Lancet 1997;350:1617-21.
- Letvin NL. Progress in the development of an HIV-1 vaccine. Science. 1998;280:1875-80.
- Nabel GJ. Challenges and opportunities for development of an AIDS vaccine. Nature 2001;410:1002-7.
- Paul WE. Can the immune response control HIV infection? Cell 1995;82:177-82.
- Igarashi T, Shibata R, Hasebe F, Ami Y, Shinohara K, Komatsu T, et al. Persistent infection with SIVmac chimeric virus having tat, rev, vpu, env, and nef of HIV type 1 in macaque monkeys. AIDS Res Hum Retroviruses 1994;10:1021-9.
- Kuwata T, Igarashi T, Ido E, Jin M, Mizuno A, Chen J, et al. Construction of human immunodeficiency virus 1/simian immunodeficiency virus strain mac chimeric viruses having vpr and/or nef of different parental origins and their in vitro and in vivo replication. J Gen Virol 1995;76:2181-91.
- Akari H, Nam KH, Mori K, Otani I, Shibata H, Adachi K, et al. Effect of SIVmac infection on peripheral blood CD4+CD8+T lymphocytes in cynomolgus macaques. Clin Immunol 1999;91:321-9.
- Clapham P, Weiss R, Dalgleish A, Exley M, Whitby D, Hogg N. Human immunodeficiency virus infection of monocytic and T-lymphocytic cells: Receptor modulation and differentiation induced by phorbol ester. Virology 1987;158:45-51.
- Igarashi T, Ami Y, Yamamoto H, Shibata R, Kuwata T, Mukai R, et al. Protection of monkeys vaccinated with vprand/or nef-defective simian immunodeficiency virus strain mac/human immunodeficiency type 1 chimeric viruses: A potential candidate live-attenuated human AIDS vaccine. J Gen Virol 1997;78:985-9.




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.