Detection of Metallo-β-lactamase (MBL) among Carbapenem-Resistant Gram-Negative Bacteria from Rectal Swabs of Cow and Cloacae Swabs of Poultry Birds
2 Department of Pharmaceutical Microbiology and Biotechnology, Faculty of Pharmaceutical Sciences, Nnamdi Azikiwe University, Awka, P.M.B 5025, Anambra State, Nigeria
3 Department of Medical Microbiology and Parasitology, Nnamdi Azikiwe University, Awka (Nnewi Campus), P.M.B 5001 Nnewi, Anambra State, Nigeria
Citation: Ejikeugwu C, et al. Detection of Metallo-β-lactamase (MBL) among Carbapenem-resistant Gram-negative Bacteria from Rectal Swabs of Cow and Cloacae Swabs of Poultry Birds. Ann Med Health Sci Res. 2017; 7: 51-56
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Background: Metallo-β-lactamases (MBLs) are carbapenem-hydrolyzing enzymes that give Gram-negative bacteria the exceptional ability to resist the antimicrobial onslaught of the carbapenems such as imipenem and meropenem. MBL-producing Gram-negative bacteria exist in the community and hospital environment and they put their use of the carbapenems at risk.
Aim: This study phenotypically evaluated the prevalence of MBL-positive bacteria from carbapenem resistant Gram-negative bacteria of abattoir and poultry origin.
Materials and methods: A total of 370 environmental samples comprising samples from abattoir tables, anal swabs of cow and cloacae swab samples of poultry birds were bacteriologically analyzed for the isolation of carbapenem Gram-negative bacteria. Antibiogram was determined by the modified Kirby-Bauer disk diffusion method and MBL production was confirmed using the modified Hodges test (MHT) technique. Nitrocefin sticks were used to screen the bacterial isolates for β-lactamase production.
Results: A total of 168 Escherichia coli, 141 Klebsiella species and 147 Pseudomonas aeruginosa isolates were recovered from the samples. More than 50% of the isolated Gram-negative bacteria were highly resistant to carbapenems, cephalosporins, aminoglycosides and fluoroquinolones. β-lactamase production was detected in E. coli (38%), P. aeruginosa (33%) and Klebsiella species isolates (29%). The E. coli isolates was resistant to imipenem (51%), meropenem (55.4%) and ertapenem (86.9%). Klebsiella species and P. aeruginosa showed resistance to imipenem, meropenem and ertapenem at the rates of 41.1%, 43.3%, 84.4%; and 66.7%, 60.5%, 61.2% respectively. MBL was phenotypically detected in 22 (39.9%) carbapenem-resistant E. coli isolates, 21 (45.7%) Klebsiella species isolates and 20 (38.9%) P. aeruginosa isolates.
Conclusion: Conclusively, this study reported the occurrence of MBLproducing E. coli, Klebsiella species and P. aeruginosa isolates from abattoir and poultry sources. The occurrence of MBL-producing bacteria in abattoir and poultry samples portends serious health implication for humans who depend on these animals for source of food; and this is due in part to the transmission of drug-resistant bacteria to human population.
Keywords
Carbapenem-resistant bacteria; Metallo-β-lactamase; Community acquired infection; Carbapenemases
Introduction
Food-producing animal’s habouring Gram-negative bacteria possessing multidrug resistant genes together with genes that provoke the production of metallo-beta-lactamases (MBLs) possess health risks to the human population. This is because bacterial pathogens positive for MBL production are resistant to the carbapenems, which are widely used to treat serious Gram-negative infections including those caused be extended spectrum β-lactamase (ESBL)-producing bacteria. One of the biggest current challenges facing the health sector across the globe especially in the area of infection control and prevention is in the adequate containment of multidrug resistant Gramnegative organisms (MDRGNOs), especially those that have been previously reported to produce MBLs. [1-4]. MBLs are carbapenem-hydrolyzing beta-lactamases which belong to molecular Class B of Ambler beta-lactamase classification, and which have the ability to hydrolyze and confer resistance to carbapenems such as imipenem, meropenem, ertapenem and doripenem. [5-7]. MBLs, which are a type of carbapenemases, are an emerging public health problem among clinically important Gram-negative organisms and environmental isolates including Pseudomonas aeruginosa, Acinetobacter baumannii and the Enterobacteriaceae. [2,3,7]. The broad spectrum activity and stability of the carbapenems to most beta-lactamase enzymes such as ESBLs makes the carbapenems an effective tool for the treatment of severe Gram-negative infections. [3,8,9]. Nevertheless, there are plethora of reports on the prevalence of MBL-producing Gram-negative bacteria from both clinical and environmental isolates around the world. [4,10-14]. The MBLs are known to confer variable range of high resistance to all betalactam antibiotics except the monobactams and their presence in clinically important Gram-negative bacteria have put the use of the carbapenems under threat. [5,7]. The MBLs belong to a group of beta-lactamases which requires divalent cations such as zinc ions as cofactors for their enzyme activity. [9]. The production of MBLs as a carbapenem resistance factors is a top arsenal of pathogenic bacteria used to make less-efficacious the therapeutic effects of the carbapenems and cephalosporins. [4,9,15]. MDRGNOs that produce MBLs in the community especially amongst animals and poultry birds are a constant source of the emergence and spread of antibiotic resistant bacteria in human population. [9,15]. These non-hospital sources of MBL-producing bacteria are of public health importance due to the possibility of transmission to humans through contact or consumption of contaminated animal and poultry products. This study evaluated phenotypically the detection of MBL among carbapenemresistant Gram-negative bacteria from abattoir and poultry sources.
Methods
Determination of sample size
Sample size determination for this study was determined by the Cochran’s formular; and a total of three hundred and seventy (370) environmental samples comprising samples from abattoir tables (n=130), anal region of cow (n=120) and the cloacae of poultry birds particularly broilers (n=120) were used in this study. The samples were collected from abattoirs and poultry farms in Abakaliki metropolis, Nigeria over a one year period (July, 2015 – June, 2016).
Isolation of bacteria
The environmental swab samples were each cultured in 5 ml double strength of nutrient broth (Oxoid, UK) and incubated overnight at 30°C. A loopful of the specimen or turbid solution was plated aseptically onto cetrimide selective agar plate(s), MacConkey agar (MAC) plates and eosin methylene blue (EMB) agar for the selective isolation of P. aeruginosa, Klebsiella species and Escherichia coli respectively. The culture plates were incubated at 30°C for 18-24 hours. P. aeruginosa isolates produces greenish pigmentation on cetrimide selective agar; E. coli also produces colonies with metallic green sheen on EMB agar and lactose-fermenting colonies on MAC; and Klebsiella species produce small, circular, elevated and mucoid colony on MAC and non-metallic sheen mucoid colonies on EMB agar. [16].
Quality control test
Quality control test to determine and confirm the isolated organisms as Escherichia coli, Pseudomonas aeruginosa and Klebsiella species was done with the reference strains: Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 10145 and Klebsiella species ATCC 700603 (Oxoid, UK); and this was based on the microscopical and morphological and/ or colonial characteristics of these control strains on culture media plates of MAC, cetrimide selective agar and EMB agar. The morphological appearances of the quality control organisms under the microscope were also used to evaluate the organisms isolated. These quality control strains were also used as standards for performing antimicrobial susceptibility testing.
Antimicrobial susceptibility testing (AST)
AST was carried out on all the recovered test bacterial isolates using the modified Kirby-Bauer disk diffusion method on Mueller-Hinton (MH) agar plates (Oxoid, UK) as was previously described, and based on the guideline of the Clinical and Laboratory Standard Institute, CLSI. [4,12,17]. This was done using single disks of: imipenem (IPM, 10 μg), meropenem (MEM, 10 μg), ertapenem (ETP, 10 μg), cefoxitin (FOX, 30 μg), ceftazidime (CAZ, 30 μg), cefotaxime (CTX, 30 μg), ceftriaxone (CRO, 30 μg), cloxacillin (OB, 500 μg), ciprofloxacin (CIP, 10 μg), aztreonam (ATM, 30 μg), ampicillin (AMP, 10 μg), nitrofurantoin (F, 10 μg), oxacillin (OX, 10 μg), ofloxacin (OFX, 10 μg), amikacin (AK, 10 μg) and gentamicin (CN, 10 μg). Antimicrobial susceptibility test results were recorded as susceptible (S), intermediate (I) and resistant (R) according to CLSI criteria. [17].
Nitrocefin test for beta-lactamase production
Beta-lactamase production by resistant test isolates was evaluated using the Nitrocefin test sticks (Oxoid, UK) as described by the method of Akinduti et al. [18].
Screening for metallo-beta-lactamase (MBL) production
To phenotypically screen for the production of MBL in the test isolates, the susceptibility of test organisms to imipenem (IPM), meropenem (MEM), and ertapenem (ETP) was evaluated as per the CLSI criteria (CLSI, 2011). Test isolates showing inhibition zone diameter (IZD) of ≤ 23 mm were considered and suspected to produce MBL enzyme; and these isolates were further tested using a phenotypic confirmation test. [6,11].
Modified Hodges Test for phenotypic detection of MBL
The modified Hodges or Cloverleaf test was performed by aseptically swabbing MH agar plates with Escherichia coli ATCC 25922 strain. The inoculated MH agar plates were allowed for about 5 min; and imipenem (10 μg) disks were aseptically placed at the center of the MH agar plates. The test bacteria (that showed reduced susceptibility to any of the carbapenems, and as adjusted to 0.5 McFarland turbidity standards) were heavily streaked from the edge of the IPM disk to the circumference of the MH agar plates. Susceptibility plates were incubated for 18-24 hrs at 30oC. The plates were macroscopically observed for indentation, and the growth of the test bacteria towards the imipenem disk. Growth of test bacteria towards the carbapenem disk is indicative of MBL production phenotypically. [6,11,13,14].
Multiple antibiotic resistance index (MARI)
Multiple antibiotic resistance indexes were determined for MBL-producing Gram-negative bacteria. The multiple antibiotic resistance index (MARI) was determined by the method of Akinjogunla and Enabulele [19]. using the formular: MARI = a/b; where ‘a’ represents the number of antibiotics to which the resistant bacteria was resistant to, and ‘b’ represents the total number of antibiotics to which the resistant bacteria has been evaluated for.
Results
In this study, environmental samples including cloacal swabs of poultry birds, anal swabs of cow and swab samples from abattoir and poultry sources were bacteriologically analyzed for the isolation of carbapenem Gram-negative bacteria that produce MBL phenotypically. Overall, E. coli was isolated from 69 swab samples out of 130 swab samples from slaughter/abattoir tables, 51 swab samples out of 120 cloacal swab samples of poultry birds, and from 48 swab samples out of 120 swab samples from the anal region of cows [Table 1]. The recovery rate of Klebsiella species isolates from the environmental swab samples including swab samples from abattoir tables, cloacal swab samples from poultry birds and anal swab samples from cows was 28.4%, 34.8%, and 36.9% respectively [Table 1]. P. aeruginosa was isolated from 56 swab samples out of 130 swab samples from abattoir/slaughter tables, 48 swab samples out of 120 cloacal swab samples, and from 43 swab samples out of 120 anal swab samples from cows. Abattoirs/slaughter houses and poultry farms are good grounds for the breeding, development and spread of antibiotic resistant bacteria including E. coli, Klebsiella species and P. aeruginosa.
| Organisms | Swabs from slaughter/abattoir benches (n = 130) n (%) | Cloacal swabs of poultry birds | Anal/rectal swabs of cow (n = 120) n (%) | Total |
|---|---|---|---|---|
| Escherichia coli | 69 (41.1) | 51 (30.4) | 48 (28.6) | 168 |
| Pseudomonas aeruginosa | 56 (38.1) | 48 (32.7) | 43 (29.3) | 147 |
| Klebsiella species | 40 (28.4) | 49 (34.8) | 52 (36.9) | 141 |
Keys: n = number of isolates; % = percentage
Table 1: Recovery rate of gram negative bacteria on bacteriological media.
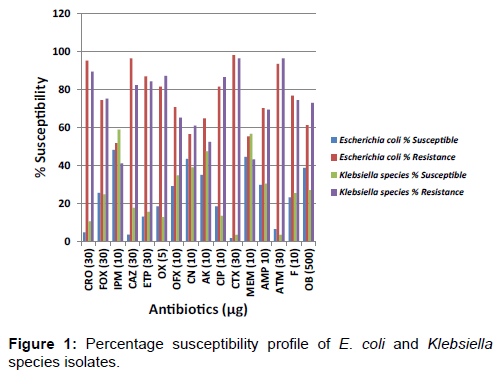
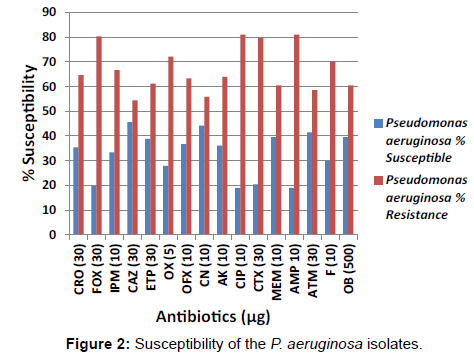
The percentage susceptibility of the Enterobacteriaceae isolates in this study including E. coli and Klebsiella species isolates to the test antibiotics is shown in Figure 1. Figure 2 shows the percentage susceptibility of the P. aeruginosa isolates to the tested antibiotics.
Very low levels of susceptibility of the E. coli isolates was observed in cefoxitin, oxacillin, ofloxacin, amikacin, ciprofloxacin, and aztreonam at a rate of 74.4%, 81.5%, 70.8%, 64.9%, 81.5% and 93.5% respectively [Figure 1]. The resistance pattern most commonly observed amongst the Klebsiella species isolates was resistance to cefotaxime (96.5%), aztreonam (96.5%), ceftriaxone (89.4%), oxacillin (87.2%), ciprofloxacin (86.5%), ceftazidime (82.3%) and cloxacillin (73.0%). The antibiotic resistance pattern of the organisms isolated in this study confirmed that more than 50% of the P. aeruginosa isolates showed high level resistance to the carbapenems including imipenem (66.7%), ertapenem (61.2%) and meropenem (60.5%). Reduced susceptibility of the P. aeruginosa isolates was also observed in cefoxitin (80.3%), cefotaxime (79.6%), ceftriaxone (64.6%) and ceftazidime (54.4%) [Figure 3].
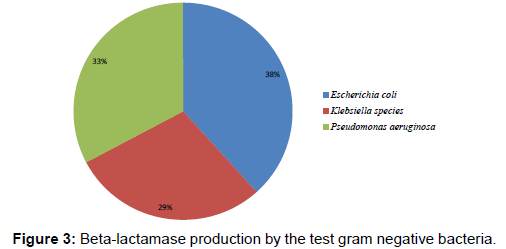
Figure 3 shows the occurrence of beta-lactamase production in the Gram-negative bacteria at varying rates. E. coli produced beta-lactamase enzyme at the rate of 38% while P. aeruginosa produced the enzyme at the rate of 33%. The rate of betalactamase production in Klebsiella species isolates was 29%.
Table 2 shows the result of the susceptibility of the bacterial isolates to the carbapenems including imipenem, meropenem and ertapenem. Out of the 168 E. coli isolates recovered in this study, a total of 87 isolates (51.8%), 93 isolates (55.4%), and 146 (86.9%) E. coli isolates were resistant to imipenem, meropenem and ertapenem respectively. A total of 58 isolates of Klebsiella species (41.1%), 61 (43.3%) Klebsiella species, and 119 (61.2%) Klebsiella species isolates were resistant to imipenem, meropenem and ertapenem respectively [Table 2]. The P. aeruginosa isolates were also resistant to imipenem (n=98, 66.7%), meropenem (n=89, 60.5%), and ertapenem (n=90, 61.5%). Notably, the P. aeruginosa isolates were more resistant to the carbapenems used in this study (imipenem, meropenem and ertapenem) than the Klebsiella species and E. coli isolates. The prevalence of MBL-producing Gramnegative bacteria is shown in Table 3. Metallo-β-lactamase (MBL) production was phenotypically detected by the modified Hodges test technique in a total of 22 E. coli isolates, 21 Klebsiella species isolates and 20 P. aeruginosa isolates. The MBL-producing E. coli, Klebsiella species and P. aeruginosa isolates had multiple antibiotics resistance index in the range of 0.6 to 0.8; and this implies that the carbapenem-resistant Gramnegative bacteria confirmed phenotypically to produce MBL in this study are multiple resistant in nature. The MBL positive E. coli, Klebsiella species and P. aeruginosa isolates were resistant to more than six (6) antibiotics out of the 16 antibiotics used in this study.
| Bacteria | Imipenem (10 μg) n (%) | Meropenem (10 μg) n (%) | Ertapenem (10 μg) n (%) |
|---|---|---|---|
| Escherichia coli (n=168) | 87 (51.8) | 93 (55.4) | 146 (86.9) |
| Klebsiella species(n=141) | 58 (41.1) | 61 (43.3) | 119 (84.4) |
| Pseudomonas aeruginosa (n=147) | 98 (66.7) | 89 (60.5) | 90 (61.2) |
Table 2 : Number of carbapenem producing gram-negative bacteria.
| Organism | Source | Number of MBL positive isolates | Percentage (%) |
|---|---|---|---|
| Escherichia coli | Abattoir | 8 | 11.6 |
| Escherichia coli | Poultry | 7 | 13.7 |
| Escherichia coli | Anal swabs of cow | 7 | 14.6 |
| Total | 22 | 39.9 | |
| Pseudomonas aeruginosa | Abattoir | 7 | 12.5 |
| Pseudomonas aeruginosa | Poultry | 8 | 14.6 |
| Pseudomonas aeruginosa | Anal swabs of cow | 6 | 18.6 |
| Total | 21 | 45.7 | |
| Klebsiella species | Abattoir | 7 | 15 |
| Klebsiella species | Poultry | 5 | 14.3 |
| Klebsiella species | Anal swab of cow | 8 | 9.6 |
| Total | 20 | 38.9 |
Table 3 : Prevalence of metallo-ß-lactamase (MBL) producing gram-negative bacteria.
Discussion
In efforts to limit the export of multidrug resistant bacteria pathogens including those that produce metallo-β-lactamase (MBL) from the hospital to the community and vice-versa, it is needful to implement sustainable monitoring and detection techniques that will enable laboratory personnel to effectively detect and report such occurrences. The effective monitoring of the development and spread of antimicrobial resistance in zoonotic pathogens including E. coli, Klebsiella species and P. aeruginosa is critical to the containment of any disease outbreak due to these drug-resistant microbes. In this present study, the production of metallo-β-lactamase (MBL) was phenotypically detected from carbapenem-resistant Gram-negative bacteria of abattoir and poultry origin. E. coli isolates, followed by P. aeruginosa isolates was the most prevalent Gram-negative bacteria isolated in this study. The increasing reports of the development and spread of Gram-negative bacteria in abattoir and poultry samples portend public health risk due in part to their possible antibiotic resistant nature and their ability to cause several bacterial infections in human populace. Akinduti et al. [18]. reported in their study that E. coli, Klebsiella species and P. aeruginosa were the most prevalent organisms isolated from environmental samples including samples from poultry farms. Beta-lactamase enzymes were phenotypically detected in the Gram-negative bacteria at varying rates. The E. coli isolates in this study produced beta-lactamase enzyme at the rate of 38% while P. aeruginosa produced the enzyme at the rate of 33%. The rate of beta-lactamase production in Klebsiella species isolates was 29%. A previous study has shown that the presence of beta-lactamase enzyme in bacteria provides opportunity for the horizontal transmission of these enzymes from one organism to another. [15]. Out of the 168 isolates of E. coli recovered from the environmental samples in this study, a total of 160 (95.2%) isolates was resistant to ceftriaxone. It was also found that 162 (96.4%) isolates of the E. coli isolates and 165 (98.2%) E. coli isolates were resistant to ceftazidime and cefotaxime. Very low levels of susceptibility of the E. coli isolates was also observed with cefoxitin, oxacillin, ofloxacin, amikacin, ciprofloxacin, and aztreonam at a rate of 74.4%, 81.5%, 70.8%, 64.9%, 81.5% and 93.5% respectively. The high levels of resistance of E. coli isolates from environmental samples (as obtainable in this study) have been reported in the Netherlands, Nigeria, and Uganda. [20-22]. Bergenholtz et al. [23]. also reported high resistance of E. coli isolates from environmental samples to antibiotics as reported in this study. Olutayo and Abimbola [24]. showed in their study that 100 E. coli isolates recovered from abattoir effluents were resistant to imipenem and meropenem [24]. Rossolini et al. [25]. reported that Enterobacteriaceae from environmental samples were highly resistant to imipenem and meropenem. In Southwest Nigeria, Ogunleye et al. [22]. reported that E. coli isolates recovered from poultry were highly resistant to imipenem and meropenem. Also in a previous study of ours, the resistance of E. coli to the carbapenems (as obtainable in this study) has also been reported [6]. Klebsiella species isolates were resistant to imipenem (41.1%), meropenem (43.3%) and ertapenem (84.4%). The resistance pattern most commonly observed amongst the Klebsiella species isolates was resistance to cefotaxime (96.5%), aztreonam (96.5%), ceftriaxone (89.4%), oxacillin (87.2%), ciprofloxacin (86.5%), ceftazidime (82.3%) and cloxacillin (73.0%). In Switzerland, the resistance of Klebsiella species to the carbapenems has also been reported as obtained in this study. [26]. The P. aeruginosa isolates recovered in this study also showed high level resistance to the carbapenems including imipenem (66.7%), ertapenem (61.2%) and meropenem (60.5%). Reduced susceptibility of the P. aeruginosa isolates was also observed in cefoxitin (80.3%), cefotaxime (79.6%), ceftriaxone (64.6%) and ceftazidime (54.4%). All the P. aeruginosa isolates also showed reduced susceptibility to oxacillin, ofloxacin, ciprofloxacin, ampicillin, aztreonam, nitrofurantoin and cloxacillin. This result is similar to results obtained by Aibinu et al. [11]. and Olutayo and Abimbola [24]. who both reported similar levels of reduced susceptibility of P. aeruginosa isolates to antibiotics in southwest Nigeria. In a recent study in southeast Nigeria, similar level of resistance amongst P. aeruginosa isolates from abattoir has also been reported. [27]. The occurrence of MBL-producing E. coli isolates in this study was 39.9%. This is similar to the work of Leung et al. [28]. who reported in Australia the occurrence of MBL-producing E. coli from environmental samples. Chakraborty et al. [29]. also reported similar prevalence of E. coli isolates positive for MBL production in India. This result is also similar to the work of Bashir et al. [30]. who recorded higher prevalence of MBLproducing E. coli isolates in their study carried out in Kashmir. The prevalence of MBL-producing E. coli isolates in this study also agreed with the work of Chouchani et al. [31]. who reported the occurrence of MBL-producing E. coli isolates (13%) in their study. The occurrence of MBL-producing Klebsiella species in this study was 38.9%. Similar rates of MBL-producing Klebsiella species have been reported in Nigeria, Asia, Europe and other parts of Africa. [6,18,29,32]. However, a similar work done in Australia showed that none of the Klebsiella species isolates recovered from environmental samples produced metallo betalactamase enzyme. [28]. In this study, P. aeruginosa isolates that produce MBL enzymes were phenotypically detected in a total of 21 (45.7%) isolates. This result is not in agreement with those reported by Abd El-Baky et al. [33]. in which 31 isolates of P. aeruginosa was phenotypically detected to produce MBL enzymes in Asia. Akinduti et al. [18]. also reported a lower rate of MBL-positive P. aeruginosa isolates (3.3%) in their study carried out in Southwest Nigeria. Shibata et al. [34]. also reported a higher occurrence rate of MBL-producing P. aeruginosa isolates in their study in which 116 P. aeruginosa isolates were discovered phenotypically to produce MBL enzymes in Japan. However, the result of MBL enzyme production amongst the P. aeruginosa isolates screened in this study is in agreement with the report of Saderi et al. [14]. who reported similar prevalence of MBL-producing P. aeruginosa isolates in their study carried out in Iran. There was no statistical difference (p value > 0.05) in the phenotypic detection of MBLs in the E. coli, Klebsiella species and P. aeruginosa isolates evaluated in this study. The effective monitoring of the development and spread of antimicrobial resistance in zoonotic pathogens especially those that produce high profile antibiotic degrading enzymes such as MBLs is critical to the containment of any disease outbreak due to these microbes. Proper sanitization, better hygienic practices and immunization should be employed in the rearing and production of food-producing animals instead of using antibiotics in order to forestall the emergence and spread of drug resistant bacteria.
Conclusion
This study reported the prevalence of MBL-producing Gramnegative bacteria from abattoir and poultry sources in Abakaliki metropolis of Ebonyi state, Nigeria. The MBL producing bacteria in this study showed high level of resistance to some commonly available antibiotics especially to those in the class carbapenems and cephalosporins. Our study indicates a possible emergence and spread of MBL-producing bacteria in abattoir and poultry sources; and this could be connoted to the irrational use of antibiotics in animal husbandry and poultry practices.
Limitations
This study was only conducted in a limited number of abattoirs and poultry farms in Abakaliki metropolis, Nigeria; and thus the findings cannot be generalized to the emergence and spread of MBL-producing bacteria in Abakaliki or Nigeria as a whole.
However, a further all-ecompassing study that incorporates molecular techniques is required to obtain sound epidemiological data as to the actual prevalence of MBL-producing bacteria in Abakaliki and Nigeria.
Acknowledgement
Our appreciation goes to members of staff of Applied Microbiology Department, Ebonyi State University, Abakaliki, Nigeria for technical support with some laboratory equipment.
Conflict of Interest
All authors disclose that there was no conflict of interest.
REFERENCES
- Toleman MA, Biedenbach D, Bennett DMC, Jones RN, Walsh TR. Italian metallo–ß–lactamases: A national problem? Report from the SENTRY Antimicrobial Surveillance Programme. Journal of Antimicrobial Chemotherapy 2005; 55:61-70.
- Tian B, Fadhil NH, Powell JE, Kwong WK, Moran NA. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honey bees. mBio 2012; 3: e00377-12
- Grundmann H, Livermore DM, Giske CG, Canton R, Rossolini GM, Campos J, et al. The CNSE Working Group. Carbapenem-non-susceptible Enterobacteriaceae in Europe: Conclusions from a meeting of national experts. Euroroundups 2010; 1-13.
- Ejikeugwu C, Duru C, Eluu S, Oguejiofor B, Ezeador C, Ogene L, et al. Isolation and phenotypic detection of metallo-beta-lactamase (MBL)-producing Klebsiella species from cow anal swabs. Global Journal of Pharmacy and Pharmaceutical Sciences 2017; 2:1-5.
- Tortola MT, Lavilla S, Miro E, Gonzalez JJ, Larrosa N, Sabate M, et al. First detection of a carbapenem–hydrolyzing metallo enzyme in two enterobacteriaceae isolates in Spain. Antimicrobial Agents and Chemotherapy 2005; 49:3492-3494.
- Ejikeugwu PC, Ugwu CM, Iroha IR, Eze P, Gugu TH, Esimone CO. Phenotypic detection of metallo-ß-lactamase enzyme in Enugu, Southeast Nigeria. American Journal of Biological, Chemical and Pharmaceutical Science 2014; 2:1-6.
- Thompson KS. Extended – spectrum ß – Lactamase, AmpC, and carbapenemase issues. Journal of Clinical Microbiology 2010; 48:1019-1025.
- Franco MRG, Caiaffa-Filho HH, Burattini MN, Rossi F. Metallo-beta-lactamases among imipenem –resistant Pseudomonas aeruginosa in a Brazilian University hospital. Clinics 2010; 65(9):825-829.
- Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-ß-lactamases: The quiet before the storm? Clinical Microbiology Review 2005; 18(2):306-325.
- Ejikeugwu C, Edeh C, Iroha I, Orji J, Eluu S, Ugbo E, et al. Antibiogram and detection of metallo-beta-lactamase (MBL) positive Escherichia coli isolates from abattoir. Nature and Science 2016; 14:65-69.
- Aibinu I, Nwanneka T, Odugbemi T. Occurrence of ESBLs and MBL in Clinical Isolates of Pseudomonas aeruginosa from Lagos, Nigeria. Journal of American Science 2007; 3:81-85.
- Javeed I, Hafeez R, Anwar MS. Antibiotic susceptibility pattern of bacterial isolates from patients admitted to a tertiary care hospital in Lahore. Biomedica 2011; 27:19-23.
- Varaiya A, Kulkarni N, Kulkarni M, Bhalekar P, Dogra J. Incidence of metallo beta – lactamase producing Pseudomonas aeruginosa in ICU patients. Indian J Med Res 2008; 127:398-402.
- Saderi H, Karimi Z, Owlia P, Bahar MA, Rad SM. Phenotypic detection of metallo–beta–lactamase producing Pseudomonas aeruginosa strains isolated from burned patients. Iranian Journal of Pathology 2008; 3:20-24.
- Bush K, Jacoby GA. Updated functional classification of ß-lactamases. Antimicrobial Agents and Chemotherapy 2010; 54:969-976.
- Cheesbrough M. District laboratory practice in tropical countries. (2nd edn). Cambridge University Press, UK. 2006; Pp. 178-187.
- Clinical Laboratory Standard Institute, CLSI. Performance standards for antimicrobial disk susceptibility test. Fifteenth informational supplement, CLSI document. 2011; M100-S15. Wayne, PA. USA.
- Akinduti PA, Ejilude O, Motayo BO, Adeyokinu AF. Emerging multidrug resistant AmpC beta-lactamase and carbapenemase enteric isolates in Abeokuta, Nigeria. Nature and Science 2012; 10: 70-74.
- Akinjogunla OJ, Enabulele IO. Virulence factors, plasmid profiling and curing analysis of multidrug resistant Staphylococcus aureus and coagulase negative Staphylococcus spp. isolated from patients with Acute Otitis Media. Journal of American Science 2010; 6:1022-1033.
- Van den Bogaard AE, London N, Driessen C, Stobberingh EE. Antibiotic resistance of feacal Escherichia coli in poultry, poultry farmers and poultry slaughterers. Journal of Antimicrobial Chemotherapy 2001; 47:763-771.
- Majalija S, Francis O, Sarah WG, Lubowa M. Antibiotic susceptibility profiles of fecal Escherichia coli isolates from dip-litter broiler chickens in Northern and Central Uganda. Veterinary Research 2010; 3:75-80.
- Ogunleye AO, Oyekunle MA, Sonibare AO. Multidrug resistant Escherichia coli isolates of poultry origin in Abeokuta, South Western Nigeria. Veterinarski Arhiv 2008; 78:501-509.
- Bergenholtz RD, Jorgensen MS, Hansen LH, Jensen LB, Hasman H. Characterization of genetic determinants of extended-spectrum cephalosporinases (ESCs) in Escherichia coli isolates from Danish and imported poultry meat. J Antimicrob Chemother 2009; 64, 207-209.
- Olutayo IF, Abimbola OA. Antibiogram of Escherichia coli and Pseudomonas strains isolated from wastewater generated by an abattoir as it journeys into a receiving river. Advances in Microbiology 2016; 6:303-309.
- Rossolini GM, Condemi MA, Pantanella F, Docquier JD, Amicosante G, Thaller MC. Metallo-ß-lactamase producers in environmental microbiota: new molecular class B enzyme in Janthino bcaterium lividum. Antimicrobial Agents and Chemotherapy 2001; 45:837-844.
- Zurfluh K, Hachler H, Nuesch-Inderbinen M, Stephan R. Characteristics of extended-spectrum beta-lactamase- and carbapenemase-producing Enterobacteriaceae isolates from rivers and lakes in Switzerland. Applied and Environmental Microbiology 2013; 79, 3021-3026.
- Iroha IR, Eromonsele OB, Moses IB, Afiukwa FN, Nwakaeze AE, Ejikeugwu PC. In vitro antibiogram of multidrug resistant bacteria isolated from Ogbete abattoir effluent in Enugu State, Nigeria. International Research Journal of Public and Environmental Health 2016; 3:1-6.
- Leung GHY, Gray TJ, Cheong ELY, Haertsch P, Gottlieb T. Persistence of related bla-IMP-4 metallo-beta-lactamase producing Enterobacteriaceae from clinical and environmental samples within a burns unit in Australia – A six-year retrospective study. Antimicrobial Resistance and Infection Control 2013; 2:1-8.
- Chakraborty D, Basu S, Das S. A study on infections caused by metallo-beta-lactamase producing gram-negative bacteria in intensive care unit patients. American Journal of infectious Diseases 2010; 6:34-39.
- Bashir D, Thokar MA, Fomda BA, Bashir G, Zahoor D, Ahmad S, et al. Detection of metallo–beta-lactamase (MBL) producing Pseudomonas aeruginosa at a tertiary care hospital in Kashmir. African Journal of Microbiology Research 2011; 5:164-172.
- Chouchani C, Marrakchi R, Ferchichi L, El-Salabi A, Walsh TR. VIM and IMP metallo-ß-lactamases and other extended-spectrum ß-lactamases in Escherichia coli and Klebsiella pneumoniae from environmental samples in a Tunisian hospital. APMIS 2011; 119:725-732.
- Deshpande P, Rodrigues C, Shetty A, Kapadia F, Hedge A, Soman R. Metallo – ß – lactamase (NDM-1) in Enterobacteriaceae: Treatment options with carbapenems compromised. Japi 2010; 58:147-149.
- Abd El-Baky RM, Abd El-Azeim NH, Gad GFM. Prevalence of extended spectrum beta lactamase, AmpC beta lactamase, and metallo beta lactamase among clinical isolates of Pseudomonas aeruginosa. Journal of Advanced Biotechnology and Bioengineering 2013; 1:22-29.
- Shibata N, Doi Y, Yamane K, Yagi T, Kurokawa H, Shibayama K, et al. PCR typing of genetic determinants for metallo-ß-lactamases and integrases carried by Gram-negative bacteria isolated in Japan, with focus on the class 3 integron. Journal of Clinical Microbiology 2003; 41:5407-5413.




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.