THE STUDY OF ATAXIA TELANGIECTASIA GENE AND CHECK POINT KINASE 2 GENE MUTATION IN A BENIGN AND MALIGNANT BREAST LESION
2 Department of Medical Laboratory Science (Histopathology Unit) college of Basic and Health Science, Achievers University, Owo, Ondo State, Nigeria
Published: 02-Nov-2022
This open-access article is distributed under the terms of the Creative Commons Attribution Non-Commercial License (CC BY-NC) (http://creativecommons.org/licenses/by-nc/4.0/), which permits reuse, distribution and reproduction of the article, provided that the original work is properly cited and the reuse is restricted to noncommercial purposes. For commercial reuse, contact reprints@pulsus.com
Abstract
Background: ATM is a Phosphatidylinositol-3-Kinase–Related Kinase(PIKKs), belonging to a family of serine/threonine kinases that also contains DNA-Dependent Protein Kinase (DNA-PK), which plays a role in the DNA Double-Strand Break (DSB) repair pathway of Non-Homologous End Joining (NHEJ), and Mammalian Target of Rapamycin (mTOR), a key autophagy regulator. ATM recognizes and amplifies the signal generated by double strand break, upon activation, CHK2 are released from chromatin and halt cell cycle progression to allow repair. The aim was to study the mutation of Ataxia telangiectasia gene and Chek2 gene in a malignant and benign breast lesion. There is a hitch in the comprehension of the concept of carcinogenesis thus, changes that occur to genes which cause cells to malfunction, causing them to grow and divide when they should not is not well understood. Materials and Methods: A total of ten formalin fixed, paraff in embedded tissue blocks, consisting of five fibroadenoma of the breast and five invasive ductal carcinoma, were retrieved from the Pathological Archives of Federal Teaching Hospital Ido-Ekiti. The tissue blocks were analysed with the use of Nucleic acid amplification techniques and the results were presented in SNPs and functional mutation through graphs and charts. Results: The results of the study showed that in ATM gene; Fibroadenoma SNPs observed, 63.7% insertion, 27.3% Transversion and 9.1% Transition. Functional mutation in fibroadenoma observed 100% Missense, 0% Nonsense and Silent. In invasive ductal carcinoma SNPs observed, 55.5% Transversion, 33.3% Insertion and 11.1% Transition. Funcional mutation in invasive ductal carcinoma observed, 49.95% Missense and Silent and 0% Nonsense. Comparison between fibroadenoma samples is 0. The comparison between invasive ductal carcinoma samples is 0. The comparison between fibroadenoma and invasive ductal carcinoma samples is 0.0048. In Chek2 gene; Fibroadenoma SNPs observed, 45% Transition, 44% Transversion and 11% Indel. Functional mutation in fibroadenoma observed 78% Missense, 22% Silent and 0% Nonsense. In invasive ductal carcinoma SNPs observed, 55% Transition, 33% Transversion and 12% Indel. Funcional mutation in invasive ductal carcinoma observed, 67% Missense, 33% Silent and 0% Nonsense. Comparison between fibroadenoma samples is 0. The comparison between invasive ductal carcinoma samples is 0. The comparison between fibroadenoma and invasive ductal carcinoma samples is 0.0333.Conclusion: Impairment of DNA damage response during the process of malignant transformation may promote and/or fuel tumorigenesis leading to accumulated genetic lesions and increased genomic instability therefore, mutations of ATM and CHK2 genes are frequently found in tumors and predispose to cancer development.
Keywords
ATM; CKEK2; DDR; DNA-PK; DSB.FA; IDC; SNPs
Introduction
ATM and ATR are Phosphatidylinositol-3-Kinase–Related Kinases (PIKKs), belonging to a family of serine/threonine kinases that also contains DNA-Dependent Protein Kinase (DNA-PK), which plays a role in the DNA DSB repair pathway of Non-Homologous End Joining (NHEJ), and Mammalian Target of Rapamycin (mTOR), a key autophagy regulator. ATM recognizes and amplifies the signal generated by DSBs, whereas ATR is activated by Single-Stranded DNA (ssDNA) generated for example by UV-induced DNA damage or interstrand DNA crosslinking (both of which lead to stalled replication forks), or by resected DSBs. In all cases these kinases are recruited to the DNA damage sites by specific DNA damage recognition proteins (i.e., DNA damage sensors), which are believed to be the Meiotic Recombination 11 Homolog A (MRE11)–RAD50–Nibrin (NBN, best known as nijmegen breakage syndrome 1, NBS1) complex (MRN complex) for ATM and Replication Protein A (RPA) complex-coated ssDNA for ATR. The principal substrates of ATM and ATR are the checkpoint effector kinases CHK2 and CHK1, respectively. Upon activation, CHK1 and CHK2 are released from chromatin and halt cell cycle progression to allow repair. In response to DSBs CHK2 triggers the G1-S checkpoint—a mechanism surveying S-phase entry—by catalyzing the activating phosphorylation of the tumor suppressor protein p53 (TP53, best known as p53), which in turn inhibits the Cyclin-Dependent Kinase 2 (CDK2)-CCNE1 (best known as cyclin E1) complex by transactivating the CDK inhibitor p2[1].
Both ATM and CHEK2 are considered as moderate-penetrance genes and are involved in DNA-double strand break repair mechanisms. In particular, the ATM protein kinase is a critical intermediary in a number of cellular responses to ionizing irradiation and possibly other stresses. In addition, its dysfunction results in abnormal checkpoint responses in multiple phases of the cell cycle. After DNA damage, ATM and DNA-dependent (DNA-PK) protein kinase activate CHK2, which in turn phosphorylates a number of downstream substrates involved in various cellular processes, including cell cycle arrest, apoptosis, DNA repair and mitosis[2].
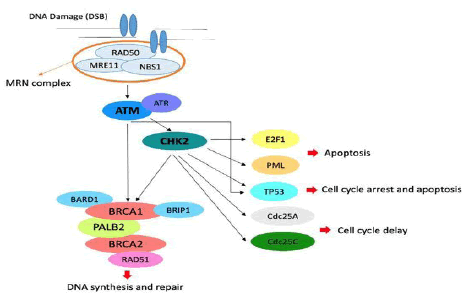
Figure 1 Role of ATM and CHK2 in the pathways of cell cycle arrest, apoptosis, DNA repair and mitosis.In particular, the MRN complex resects DNA at the Double-Strand Break (DSB) and recruits ATM that phosphorylates CHK2 and recruits the BRCA complex[2].
Material and Methods
Tissue blocks
For this retrospective study, a total of ten formalin fixed paraffin embedded tissue blocks, consisting of five fibroadenoma and five invasive ductal carcinoma of the breast were retrieved from the Pathological Archives of Federal Teaching Hospital Ido-Ekiti.
DNA extraction
The following protocols were adopted for extracting good quality DNA from paraffin embedded tissue. Each tissue was ground in sterile mortal and pistil with 500 µl of extraction buffer before being poured into a sterilized Eppendorf tube and 33 µl of 20% Sodium Dodecyl Sulphate (SDS) was added, vortexed briefly, and incubated in a water bath at 65℃ for 10 minutes. 10 µl of 5M potassium acetate was then added at room temperature, vortexed, and centrifuged at 10000 g for 10 minutes. The supernatant was collected in another Eppendorf tube, and 300l of cold iso-propanol was added. The mixture was gently mixed, and it was kept at -20℃ for 60 minutes. The DNA was sedimented by centrifugation at 13000 g for 10 minutes, after which the supernatant was gently decanted and the pellet was not disturbed. After washing the DNA pellet with 500 µl of 70% ethanol, it was centrifuged at 10000 g for 10 minutes. Ethanol was decanted, and the DNA was air-dried at room temperature until there was no trace of ethanol left in the tube. To preserve and suspend the DNA, the pellet was resuspended in 5 µl of Tris EDTA buffer [3].
Polymerase chain reaction
Sequencing preparation cocktail for all PCR consisted of 10 µl of 5x GoTaq colorless reaction, 3 µl of 25 mM MgCl2, 1 µl of 10 mM of dNTPs mix, 1 µl of 10 pmol each primers and 0.3units of Taq DNA polymerase (Promega, USA) made up to 35 µl with sterile distilled water 15 μl DNA template. PCR was carried out in a GeneAmp 9700 PCR System Thermalcycler (Applied Biosystem Inc., USA) with a PCR profile for each primer presented in the table below[4].
Integrity test
To validate the amplification, the integrity of the amplified DNA of about 1.5 Mb gene fragment was tested on an Agarose gel of about 1%. The buffer (1XTAE buffer) was prepared and then used to make the 1.5% agarose gel. The suspension was microwaved for 5 minutes. The molten agarose was allowed to cool to 60℃ before being stained with 3 µl of 0.5 g/ml ethidium bromide (which absorbs invisible UV light and transmits the energy as visible orange light). A comb was inserted into the slots of the casting tray, and molten agarose was poured into the tray. The wells were formed by allowing the gel to solidify for 20 minutes. The 1XTAE buffer was poured into the gel tank, barely submerging the gel. After loading the 100bp DNA ladder into well 1, two microliters (2 µl) of 10X blue gel loading dye (which gives the samples color and density to make it easy to load into the wells and monitor the progress of the gel) was added to 4µl of each PCR product and loaded into the wells. The gel was electrophoresed at 120 V for 45 minutes, photographed, and then visualized using ultraviolet trans-illumination. The sizes of the PCR products were estimated by comparing them to the mobility of a 100 bp molecular weight ladder that was run in the gel alongside the experimental samples[3].
Purification of amplified product
After gel integrity, the amplified fragments were ethanol purified in order to remove the PCR reagents. Briefly, 7.6 µl of Na acetate 3 M and 240 µl of 95% ethanol were added to each about 40µl PCR amplified product in a new sterile 1.5 µl tube eppendorf, mix thoroughly by vortexing and keep at -20℃ for at least 30 min. Centrifugation for 10 min at 13000 g and 4℃ followed by removal of supernatant (invert tube on trash once) after which the pellet were washed by adding 150 µl of 70% ethanol and mix then centrifuge for 15 min at 7500 g and 4℃. Again remove all supernatant (invert tube on trash) and invert tube on paper tissue and let it dry in the fume hood at room temperature for 10 min-15 min. Then resuspend with 20 µl of sterile distilled water and kept in -20℃ prior to sequencing. The purified fragment was checked on a 1.5% A garose gel ran on a voltage of 110V for about 1hr as previous, to confirm the presence of the purified product and quantified using a nanodrop of model 2000 from thermo scientific[5].
Sequencing
The amplified fragments were sequenced using a Genetic Analyzer 3130xl sequencer from Applied Biosystems using manufacturers’ manual while the sequencing kit used was that of BigDye terminator v3.1 cycle sequencing kit. Bio- Edit software and MEGA 6 were used for all genetic analysis[6].
Data analysis
The use of charts, tables and graphs was employed in the analysis of the results obtained from this study.
Results
In the Table 1 analysis of the results obtained from this study.
| Table 1: PCR profile was employed in the analysis of the results obtained from this study. | |||||||
|---|---|---|---|---|---|---|---|
| Description | Location | Gene type | % occurrence | Specimen | Mutation type | Mutation type Description | |
| Â Â C | 169(2:3) | ATM | 2(5.3) | FA(Fib) | Insertion | Leucine deletes | |
| Gene | |||||||
| Â Â G | 199(1:1) | ATM | 5(13.2) | FA(Fib) | Insertion | Insertion of Valine | |
| Gene | |||||||
| C:A | 81(1:4) | ATM | 3(7.9) | FA(Fib) | Transversion | Missense mutation changing histidine to Glutamine | |
| Gene | |||||||
| T:C | 137(9:1) | ATM | 1(2.6) | FA(Fib) | Transition | Missense mutation changing leucine to proline | |
| Gene | |||||||
| Â Â C | 169(2:3) | ATM | 4(10.5) | IDC | Insertion | Leucine deletes | |
| Gene | |||||||
| Â Â G | 199(1:1) | ATM | 5(13.2) | IDC | Insertion | Insertion of Valine | |
| Gene | |||||||
| C:A | 81(1:4) | ATM | 5(13.2) | IDC | Transversion | Missense mutation changing histidine to Glutamine | |
| Gene | |||||||
| T:A | 118(3:2) | ATM | 4(10.5) | IDC | Transversion | Missense mutation changing phenylalanine to isoleucine | |
| Gene | |||||||
| T:G | 301(4:1) | ATM | 2(5.3) | IDC | Transversion | Silent mutation retaining proline | |
| Gene | |||||||
| T:G | 375(3:2) | ATM | 4(10.5) | IDC | Transversion | Silent mutation retaining Valine | |
| Gene | |||||||
| T:C | 279(3:2) | ATM | 3(7.9) | IDC | Transition | Silent mutation retaining glycine | |
| Gene | |||||||
| T | 92(9:1) | CHEK2 | 1(0.84) | IDC | Indel | Missense mutation deleting leucine | |
| C | 93(7:3) | CHEK2 | 4(3.38) | FIB | Indel | Missense mutation deleting leucine | |
| A | 123(4:3) | CHEK2 | 6(5.08) | IDC | Indel | Missense mutation inserting Alanine | |
| T | 293(9:1) | CHEK2 | 1(0.84) | IDC | Indel | Missense mutation deleting phenylalaine | |
| G | 294(9:1) | CHEK2 | 1(0.84) | FIB | Indel | Missense mutation deleting Glycine | |
| G | 630(3:2) | CHEK2 | 6(5.08) | FIB | Indel | Missense mutation deleting Glycine | |
| T:A | 206(4:1) | CHEK2 | 2(1.69) | IDC | Transversion | Missense mutation changing Arginine to Cysteine | |
| T:A | 302(3:2) | CHEK2 | 4(3.38) | IDC | Transversion | Missense mutation changing Serine to Arginine | |
| T:A | 330(9:1) | CHEK2 | 1(0.84) | IDC | Transversion | Missense mutation changing Serine to Arginine | |
| T:A | 444(1:10 | CHEK2 | 4(3.38) | IDC | Transversion | Misense mutation changing Leucine to Arginine | |
| T:A | 456(4:1) | CHEK2 | 2(1.69) | IDC | Transversion | Missense mutation changing Phenlyalaline to Leucine | |
| T:A | 470(4:1) | CHEK2 | 2(1.69) | FIB | Transversion | Missense mutation changing Valine to Aspartate | |
| A:T | 587(9:1) | CHEK2 | 1(0.84) | IDC | Transversion | Missense mutation changing Isoleucine to Phenyalaline | |
| T:A | 655(9:1) | CHEK2 | 1(0.84) | FIB | Transversion | Misense mutation changing Phenylalaline to Isoleucine | |
| C:A | 465(2:3) | CHEK2 | 6(5.08) | IDC | Transversion | Missense mutation changing Glutamate to Aspartate | |
| C:A | 553(3:2) | CHEK2 | 4(3.38) | IDC | Transversion | Silent mutation retaining Tryosine | |
| C:A | 575(4:1) | CHEK2 | 2(1.69) | IDC | Transversion | Missense mutation changing Alaline to Aspartate | |
| G:T | 306(4:1) | CHEK2 | 2(1.69) | FIB | Transversion | Silent mutation retaining Leucine | |
| G:T | 347(7:3) | CHEK2 | 3(2.54) | IDC | Transversion | Missense mutation changing Glycine to Cystine | |
| G:T | 390(7:3) | CHEK2 | 3(2.54) | FIB | Transversion | Silent mutation retaining Leucine | |
| G:T | 439(4:1) | CHEK2 | 2(1.69) | FIB | Transversion | Missense mutation changing Glycine to Cystine | |
| G:T | 471(4:1) | CHEK2 | 4(3.38) | IDC | Transversion | Missense mutation changing Glycine to Cystine | |
| T:G | 581(4:1) | CHEK2 | 2(1.69) | FIB | Transversion | Missense mutation changing Glycine to Cystine | |
| G:C | 142(4:1) | CHEK2 | 2(1.69) | FIB | Transversion | Missense mutation changing Glycine to Cystine | |
| C:G | 145(7:3) | CHEK2 | 3(2.54) | IDC | Transversion | Missense mutation changing Glycine to Cystine | |
| G:A | 84(3:2) | CHEK2 | 4(3.38) | FIB | Transistion | Silent mutation retaining serine | |
| A:G | 175(9:1) | CHEK2 | 1(0.84) | FIB | Transistion | Missense mutation chaninging Asparate to Glycerine | |
| G:A | 363(7:3) | CHEK2 | 3(2.54) | IDC | Transistion | Silent mutation retaining Theonine | |
| A:G | 457(4:1) | CHEK2 | 2(1.69) | IDC | Transistion | Missense mutation changing serine to Arginine | |
| G:A | 546(4:1) | CHEK2 | 2(1.69) | IDC | Transistion | Missense mutation changing tryptophan to stop code | |
| G:A | 554(4:1) | CHEK2 | 2(1.69) | IDC | Transistion | Missense mutation changing Arginine to Glutamine | |
| G:A | 678(3:2) | CHEK2 | 3(2.54) | IDC | Transistion | Silent mutation retaining lycine | |
| C:T | 111(4:1) | CHEK2 | 2(1.69) | IDC | Transistion | Silent mutation retaining cysteine | |
| T:C | 122(7:3) | CHEK2 | 3(2.54) | FIB | Transistion | Â Missense mutation changing leucine to serline | |
| C;T | 124(9:1) | CHEK2 | 1(0.84) | IDC | Transistion | Missense mutationchanging leucine to phenylanine | |
| C:T | 143(4:1) | CHEK2 | 2(1.69) | FIB | Transistion | Missense mutation changing cystine to serine | |
| T:C | 276(4:1) | CHEK2 | 2(1.69) | FIB | Transistion | Silent mutation retaining isoleucine | |
| T:C | 305(7:3) | CHEK2 | 3(2.54) | IDC | Transistion | Missense mutation changing valine to leucine | |
| C:T | 380(3:2) | CHEK2 | 3(2.54) | IDC | Transistion | Missense mutation changing Alanine to threonine | |
| T:C | 434(3:2) | CHEK2 | 4(3.38) | FIB | Transistion | Silent mutation retaining proline | |
| T:C | 438(9:1) | CHEK2 | 1(0.54) | IDC | Transistion | Silent mutation retaining serine | |
| T:C | 504(7:3) | CHEK2 | 3(2.54) | FIB | Transistion | Missense mutation changing Arginine to stop code | |
| C:T | 514(3:2) | CHEK2 | 4(3.38) | IDC | Transistion | Silent mutation retaining Alanine | |
| C:T | 519(7:) | CHEK2 | 3(2.54) | IDC | Transistion | Missense mutation changing cysteine to Arginine | |
| C:T | 523(4:1) | CHEK2 | 2(1.69) | IDC | Transistion | Missense mutation changing proline to leucine | |
| C:T | 566(4:1) | CHEK2 | 2(1.69) | FIB | Transistion | Silent mutation retaining serine | |
| C:T | 609(4:1) | CHEK2 | 2(1.69) | IDC | Transistion | Silent mutation retaining serine | |
| C:T | 664(9:1) | CHEK2 | 1(0.54) | IDC | Transistion | Silent mutation retaining Glycine | |
| C:T | 669(9:1) | CHEK2 | 1(0.54) | IDC | Transistion | Sillent mutation retaining Alanine | |
| G:A:T | 423(6:1:3) | CHEK2 | 4(3.38) | IDC | Transistion | Silent mutation retaining Threonine | |
| G:A:T | 603(6:3:1) | CHEK2 | 2(1.69) | IDC | Transistion | Silent mutation retaining Threonine | |
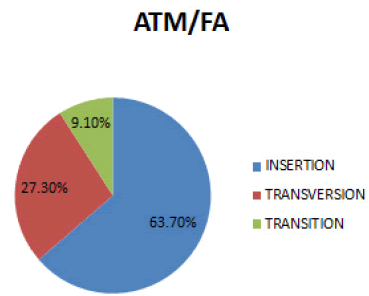
In figure 2 Insertion is the most prevalent mutation (63.7%), followed by Transversion (27.3%) and then Transition (9.1%).
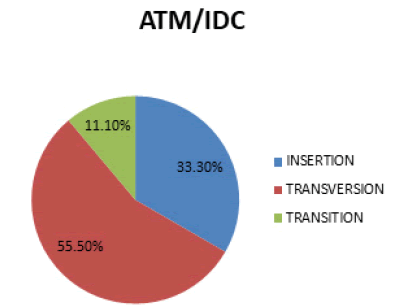
In figure 3 Transversion is the most prevalent mutation (55.5%), followed by Insertion (33.3%) and then Transition (11.1%).
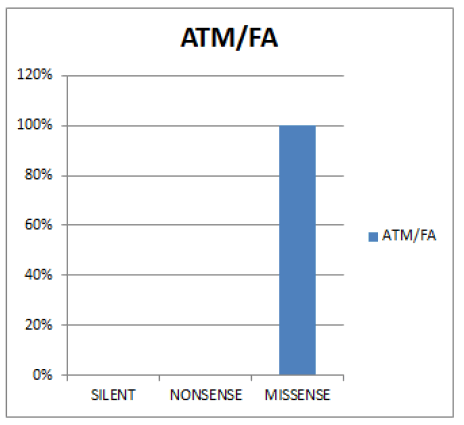
In figure 4 Missense is the most prevalent (100%) while Silent and Nonsense are (0%).
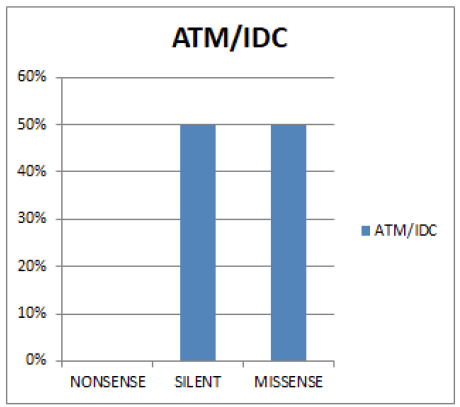
In figure 5 Both Silent and Missense mutation have (49.95%) and Nonsense (0%).
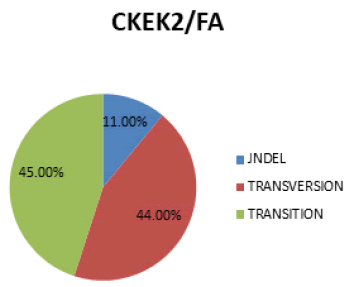
In figure 6 Transition is the most prevalent mutation (45%), followed by Transversion (44%) and then Indel (11%).
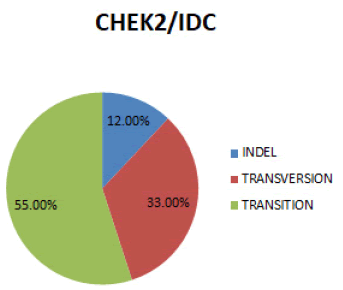
In figure 7 Transition is the most prevalent mutation (55%), followed by Transversion (33%) and then Indel (12%).
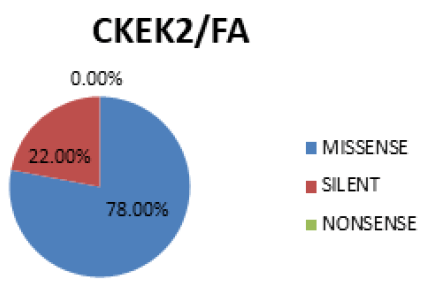
In figure 8 Missense is the most prevalent mutation (78%), followed by Silent (22%). No occurrence of Nonsence mutation (0%).
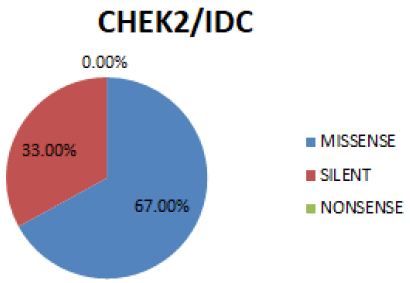
In figure 9 Missense is the most prevalent mutation (67%), followed by Silent (33%). No occurrence of Nonsense mutation (0%).
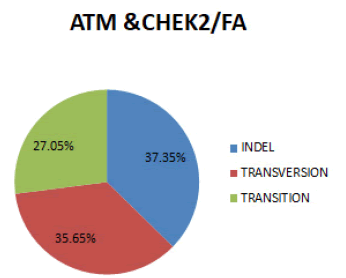
In figure 10 In both genes combined, the most prevalent SNPs for Fibroadenoma is Indel (37.35%), followed by Transversion (35.65%) and Transition (27.05%).
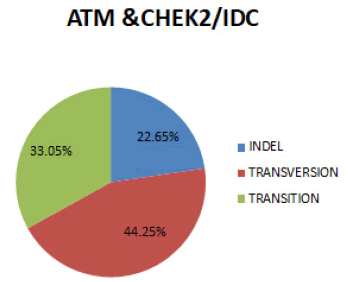
In figure 11 In both genes combined, the most prevalent SNPs for Invasive ductal carcinoma is Transversion (44.25%), followed by Transition (35.65%) and Indel (22.65%).
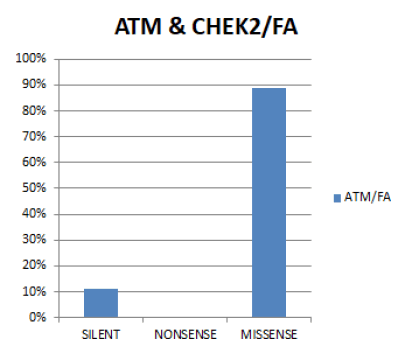
In figure 12 In both genes combined, the most prevalent Functional mutation in fibroadenoma is Missense (89%), followed by Silent (11%). No case of Nonsense mutation was seen in both genes.
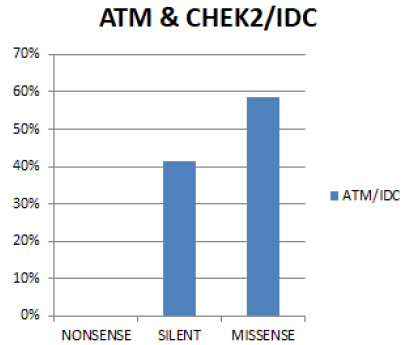
In figure 13 In both genes combined, the most prevalent Functional mutation in Invasive ductal carcinoma is Missense (58.5%), followed by Silent (41.5%). No case of Nonsense mutation was seen in both genes.
Discussion
Mutations that occurred in this study appeared to have caused abnormal cell proliferation. For ATM gene, Insertion is the most prevalent SNPs mutation in fibroadenoma . Insertion of a larger sequence of nucleotide base pairs into a chromosome may occur when there is an unequal crossover during meiosis. Insertions can be particularly hazardous if they occur in an exon, the amino acid coding region of a gene[7]. The second most common type of SNPs mutation in Fibroadenoma is Transversion. This Transversion mutation may lead to no change in the protein sequence (known as silent mutations), change the amino acid sequence (known as missense mutations), or create what is known as a stop codon (known as a nonsense mutation)[8]. Because Transversion cause larger changes in the shape of a DNA backbone, transversion would have larger impacts on transcription factors binding and gene expression[9].The least occurrence of SNPs mutation in fibroadenoma was seen in Transition. Transitions could result from oxidative deamination and tautomerization. Although there are twice as many possible transversions, transitions appear more often in genomes, possibly due to the molecular mechanisms that generate them[10]. Transitions are less likely to result in amino acid substitutions (due to "wobble"), and are therefore more likely to persist as silent substitutions[11].
In Invasive ductal carcinoma, the most prevalent SNPs mutation is Transversion. The transversion can be spontaneous, or it can be caused by ionizing radiation or alkylating agents[12]. A transversion ususlly has more pronounced effect than a transition because the third nucleotide codon position of the DNA, which to a large extent is responsible for the degeneracy of the code, is more tolerant of transition than a transversion[13]. The second most common SNPs mutation in Invasive ductal carcinoma is Insertion. Insertion indicates the removal of one segment from one chromosome and then inserting into a broken region of a non-homologous chromosome. When one or more nucleotides is inserted into the genetic sequence, the reading frame is disrupted during transcription, and eventually, translation. This may result in an altered sequence of amino acids, extra amino acid in a chain, or premature termination. Hence, the newly synthesized protein could be abnormally short, abnormally long, and/or contain the wrong amino acids[14]. Transition is the least common SNPs mutation in invasive ductal carcinoma. Transition more likely persist as silent substitution. More of transversion was seen compared to transition because transversion has more regulatory effect than transition. Transversions are more likely to alter the amino acid sequence of proteins than transitions. Whether the two different types of mutations have different effects in non-protein-coding sequences remains unknown[15].
In the functional mutation for Fibroadenoma, only Missense mutation was recorded. Missense mutation results in codon change which lead to the production of new protein, this resulting protein may fail to work properly.
In the functional mutation for Invasive ductal carcinoma, there is also a high occurrence of Missense and this indicates the instability and rapid degradation of genes. Genes may fail to localize in its proper intracellular position. A greater number of transversion led to missense mutation in this study. Silent mutation is also prevalent in invasive ductal carcinoma and this result in a new codon that codes for the same amino acid which affects protein translation efficiency and protein folding function[16].
For CHEK2 gene, in the SNPs (Single Nucleotide Polymorphisms) for FA (Fibroadenoma), the most prevalent mutation is Transversion and this transversion mutation may lead to no change in the protein sequence, changes the amino acid and create what is known as a stop codon because a greater number of transversion leads to a missense mutation and tranversions are more detrimental because they are more likely to cause substitutions that can alter biochemical properties of amino acid. The second most prevalence type of the SNPs is Fiboadenoma is Transition, this can also be dangerous, because it can cause alteration or loss of amino acid and the Indel is the least of the SNPs in Fibroadenoma, which is also known as insertion or deletion, the indels are less likely to disrupt folding in the core of protein and it can be used as genetic biomarkers[17].
For SNPs in IDC (Invasive Ductal Adenocarcinoma), Transition mutation is the most prevelant and this can leads to alteration in amino acid sequences, the second most prevalence is the transversion mutation and the least is the indel mutation. More of transition is recorded because the transition has more regulatory effect than the tranversion. For Functional Mutation in Fibroadenoma the most prevalent mutation is the missense mutation and this results in codon change which lead to the production of new protein and the protein many fail effectively and the second most prevalent functional mutation is silent mutation, also known as a neutral mutation because the nucleotide change does not change the amino acid being translated and the least is nonsense mutation which is also known as stop codon and is Zero because it occurs when a premature nonsense or stop codon is introduced in the DNA sequence [18].
In the functional mutation for Invasive ductal carcinoma, high missense mutation was recorded and it signifies the instability and rapid degradation of genes. A higher number of transversion led to missense mutation in this study. Silent mutation is the second prevalent mutation in the Invasive ductal carcinoma and may lead to new codon for amino acid which affects the protein translation efficiency and protein folding function[19].
Conclusion
Impairment of DNA damage response during the process of malignant transformation may promote and/or fuel tumorigenesis leading to accumulated genetic lesions and increased genomic instability therefore, mutations of ATM and CHK2 genes are frequently found in tumors and predispose to cancer development.
References
- Shaik A, Kirubakaran S. Evolution of PIKK family kinase inhibitors: A new age cancer therapeutics. Frontiers in Bioscience-Landmark. 2020; 25:1510-1537.
[Crossref] [Google Scholar] [Indexed]
- Toss A, Tenedini E, Piombino C, Venturelli M, Marchi I, Gasparini E, et al. Clinicopathologic profile of breast cancer in germline ATM and CHEK2 mutation carriers. Genes. 2021; 12:616.
[Crossref] [Google Scholar] [Indexed]
- Odeyemi AT, Ayantola KJ, Peter S. Molecular characterization of bacterial isolates and physicochemical assessment of well water samples from hostels at Osekita, Iworoko-Ekiti, Ekiti State. American Journal of Microbiological Research. 2018; 6:22-32.
- Batista FM, Stapleton T, Lowther JA, Fonseca VG, Shaw R, Pond C, et al. Whole genome sequencing of hepatitis a virus using a pcr-free single-molecule nanopore sequencing approach. Front Microbiol. 2020; 11:874.
[Crossref] [Google Scholar] [Indexed]
- https://www.fisheriesjournal.com/archives/2021/vol9issue3/PartB/9-2-29-327.pdf
- Njoku KL, Asunmo MO, Ude EO, Adesuyi AA, Oyelami AO. The molecular study of microbial and functional diversity of resistant microbes in heavy metal contaminated soil. Environmental Technology & InOctation.2020;17:100606.
- Merrick L, Campbell A, Muenchrath D, ISU SZF. Mutations and variation. 2021.
- Sharma Y, Miladi M, Dukare S, Boulay K, Caudron-Herger M, Groß M, et al. A pan-cancer analysis of synonymous mutations. Nature communications. 2019;10:1-14.
[Crossref] [Google Scholar] [Indexed]
- Guo C, McDowell IC, Nodzenski M, Scholtens DM, Allen AS, Lowe WL, et al. Transversions have larger regulatory effects than transitions. BMC genomics. 2017;18:394.
[Crossref] [Google Scholar] [Indexed]
- Kay J, Thadhani E, Samson L, Engelward B. Inflammation-induced DNA damage, mutations and cancer. DNA repair.2019; 83:102673.
[Crossref] [Google Scholar] [Indexed]
- https://www.proquest.com/openview/1996b1a906767d29695f811512387bd9/1?pqorigsite=gscholar&cbl=2042720.
- Ullmann R, Becker BV, Rothmiller S, Schmidt A, Thiermann H, Kaatsch HL, et al. Genomic adaption and mutational patterns in a HaCaT subline resistant to alkylating agents and ionizing radiation. Int J Mol Sci. 2021; 22: 1146.
[Crossref] [Google Scholar] [Indexed]
- Nemzer LR. Shannon information entropy in the canonical genetic code. J Theor Biol. 2017; 415:158-170.
[Crossref] [Google Scholar] [Indexed]
- Lee M, Soltanian HT. Breast fibroadenomas in adolescents: Current perspectives. Adolesc Health Med Ther. 2021; 6:159.
[Crossref] [Google Scholar] [Indexed]
- Kakatkar AS, Das A, Shashidhar R. Ciprofloxacin induced antibiotic resistance in Salmonella Typhimurium mutants and genome analysis. Archives of Microbiology. 2021;203:6131-6142.
[Crossref] [Google Scholar] [Indexed]
- Liu Y. (2020). A code within the genetic code: Codon usage regulates co-translational protein folding.Cell Commun Signal. 2020;18:1-9.
[Crossref] [Google Scholar] [Indexed]
- Mullaney JM, Mills RE, Pittard WS, Devine SE. Small insertions and deletions (INDEL) in human genomes. Human Molecular Genetics. 2010;19(R2):R131-6.
[Crossref] [Google Scholar] [Indexed]
- Wooster R, Renwick K, Tavtigian SV, Simard J, Rommes J. Linkage of of early onset familial breast cancer to chromosome. 2019.
- Samatova E, Daberger J, Liutkute M, Rodnina MV. Translational control by ribosome pausing in bacteria: how a non-uniform pace of translation affects protein production and folding. Frontiers in microbiology. 2021; 11:619430.
[Crossref] [Google Scholar] [Indexed]




 The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.
The Annals of Medical and Health Sciences Research is a monthly multidisciplinary medical journal.